Fig. 3.
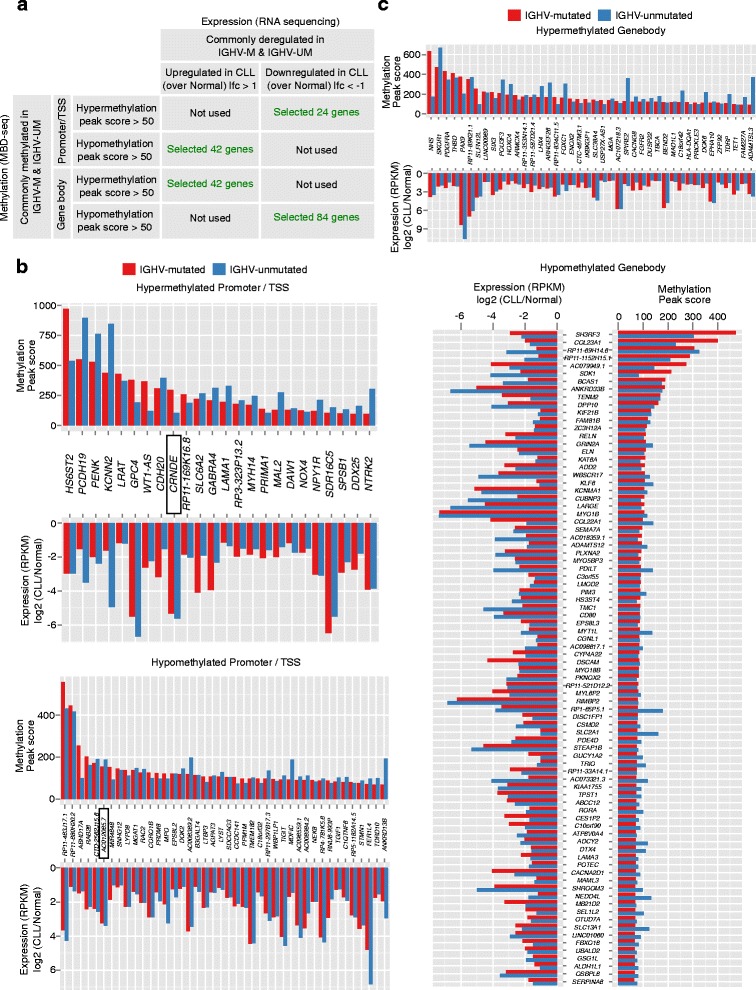
Regulation of cllDMGs by the distribution of methylation on gene structure and the gene expression patterns associated with methylation. a Table showing the selection of candidate genes depending on correlation between location of methylation on gene structure (promoter or gene body methylation in MBD-seq) and their pattern of gene expression (up or downregulated in RNA-seq, log2fold-change). The “selected number of genes” in green represents the candidate genes (cllDMGs) considered for further investigation. The two selected genes (CRNDE and AC012065.7) for further investigation from two categories were highlighted in bar graphs (b and c) with a rectangle. b The top and bottom bar graphs represent the list of selected cllDMGs from promoter-hypermethylated-downregulated and promoter-hypomethylated-upregulated patterns, respectively. c The top and bottom bar graphs show the list of selected cllDMGs from gene body-hypermethylated-upregulated and gene body-hypomethylated-downregulated patterns, respectively
