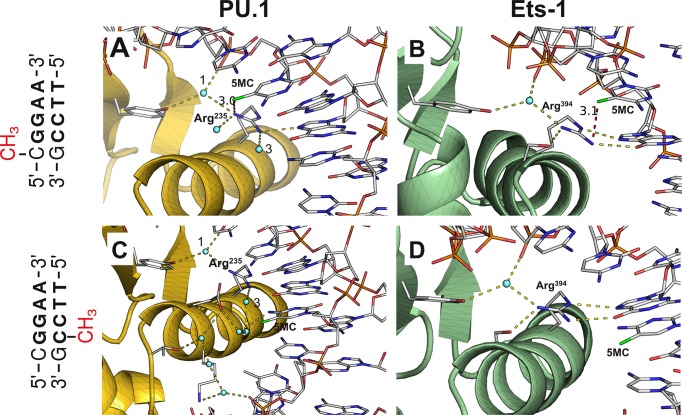
Figure 3.
Structural basis of differential inhibition of PU.1 and Ets-1 binding at hemi-methylated site-specific DNA. The high-affinity DNA sequences in co-crystal structures of the ETS domain of PU.1 (PDB: 1PUE) and Ets-1 (1K79) were mutated to methylated SC1 without modification of backbone coordinates. The protein/DNA interface near the methylated CpG dinucleotide is shown. The peptide backbones of PU.1 (Panels A and B) and Ets-1 (Panels C and D) are shown in gold and green, respectively, and only protein sidechains involved in the indicated interactions are rendered for clarity. Hydrogen bonds (2.8 ± 0.6 Å) emanating from the essential arginine (Arg235 in PU.1, Arg394 in Ets-1) at the recognition helix are shown in yellow. The closest heavy atom from the 5-methyl (colored green) of 5-methylcytosine (5MC) is joined by a red dash with distance in Å. For the PU.1/DNA complex, only the ordered waters of first contact are shown in Panel A, and only one hydration ‘train’ is shown in Panel C; note the same ordered waters marked ‘1’ and ‘3’ in the two panels. For the Ets-1/DNA complex, only ordered water participating in H-bonding with the protein is shown in Panels B and D.