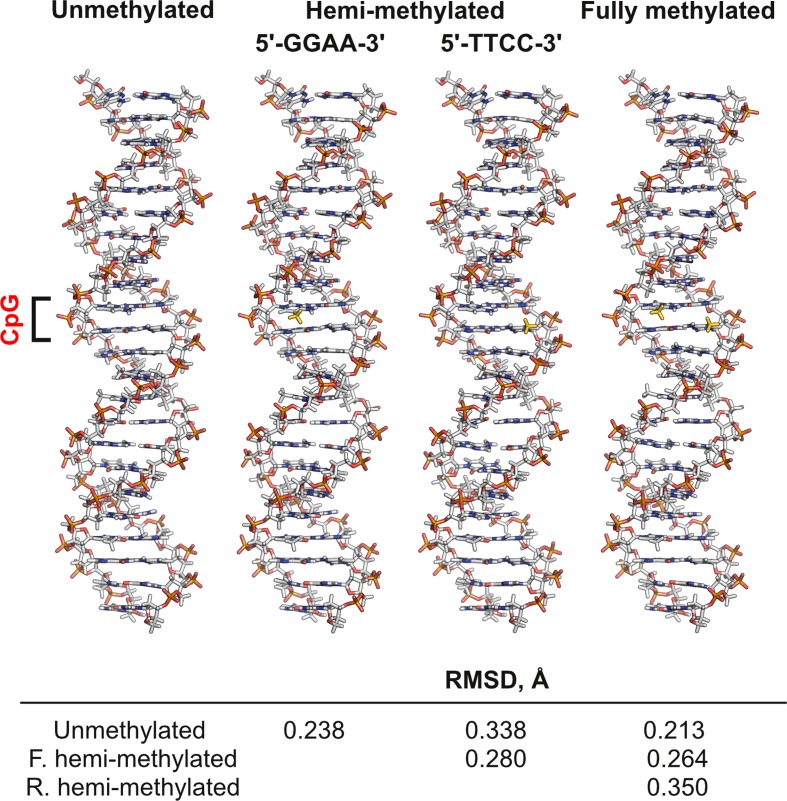
Figure 4.
Simulated structures of unmethylated and methylated cognate ETS-binding sites. A duplex DNA harboring the same SC1 sequence as used in the titration experiments (5′-CGGCCAAGCGGAAGTGAGTGCC-3′) at various methylation states was simulated by all-atom energy minimization at physiological ionic strength using the CHARMM36 force field as described in Materials and Methods. The RMSDs of pairwise structural alignments are tabulated. The structures are convergent as judged by the change in total potential energies (Figure S4, Supplemental Data). The CpG dinucleotide is bracketed and 5-methyl substituents are colored in gold.