Figure 1.
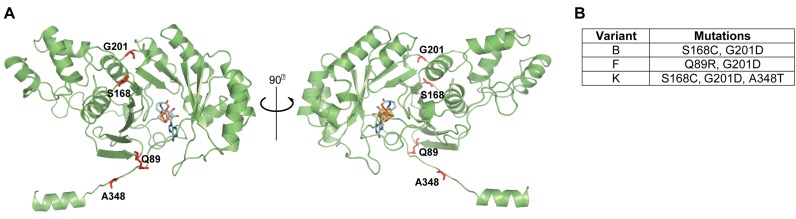
Evolved variants of Bsub_RlmN contain specific mutations distal from the active site. (A) A homology model of the structure of Bsub_RlmN was generated by I-TASSER. Shown in sticks are the iron-sulfur center (gold) and the SAM cofactor (light blue) borrowed from the aligned template structure (PDB 3RFA). The most dominant mutations present in the evolved variants are shown as red sticks. Among the mutated residues present in the Bsub_RlmN variants studied herein, only A348, part of the C-terminal region that undergoes conformational change upon substrate binding, is a part of the active site, while other residues are located >12 Å from SAM's methyl group (38). (B) Mutations present in the 3 evolved Bsub_RlmN variants analyzed in this study.