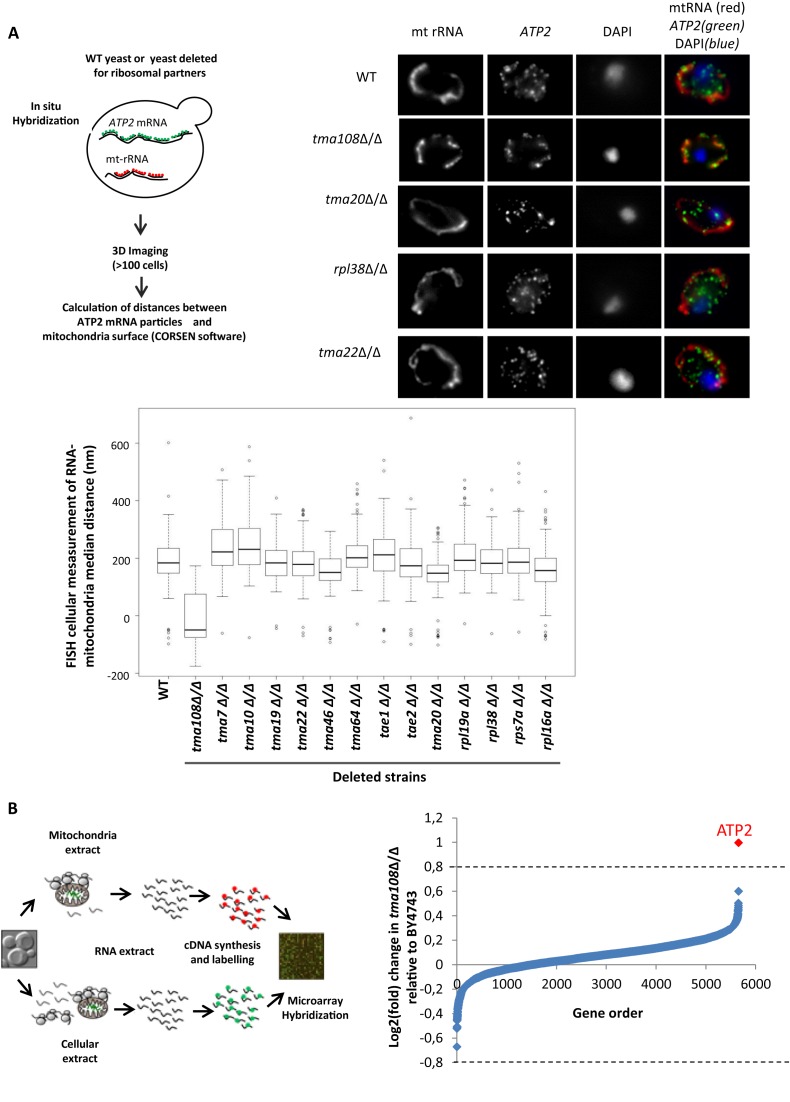
Figure 1.
The Translation Machinery-Associated factor Tma108 impacts ATP2 mRNA localization to mitochondria. (A) The impact of different translation machinery-associated factors on ATP2 mRNA localization. Experimental approach (top left). Fluorescent in situ experiments were performed in different mutants using probes to detect ATP2 mRNA and mitochondrial rRNA as a marker for mitochondria position. Typical FISH images (top right) show the impact of TMA108 deletion on ATP2 mRNA localization. In BY4743 strain, the ATP2 mRNA is partially localized to the mitochondria surface. TMA108 deletion led to a drastic increase in ATP2 mRNA localization to the mitochondria: most of the green spots co-localized with the mitochondrial red signal. ATP2 mRNA localization is not perturbed by TMA20, RPL38 or TMA22 deletions. The boxplots (bottom) represent the distributions of the median ATP2 mRNA-mitochondria distances in the analyzed cell populations. Similar FISH analyses were performed on a control mRNA, ATP16, and no significant change in mRNA localization was observed (data not shown). (B) Genome-wide analysis of the impact of Tma108 on the localization of mRNA to mitochondria. Experimental scheme (left) for the analysis of mRNA localization to mitochondria: total mRNA and mRNA associated with the mitochondrial fraction were isolated, reverse transcribed, and labeled for subsequent microarray analysis. Comparative analysis of the mRNA associated with the mitochondrial fraction in BY4743 and TMA108 deleted strain (right). The log2 fold changes in TMA108Δ/Δ relative to BY4743 are plotted: the values for each gene are the means of three biological replicates (see Supplemental Table S5). ATP2 is the single mRNA detected by LIMMA with a significant change in mRNA enrichment factor (P-value = 3.7 × 10−3).