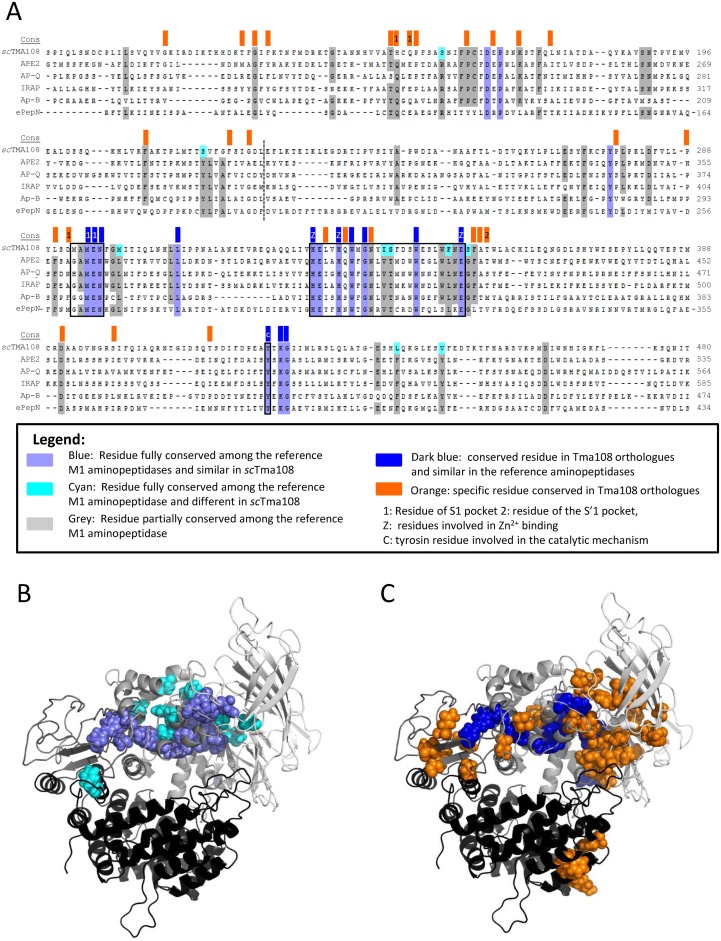
Figure 4.
Tma108, a new protein of the M1 aminopeptidase family. (A) S. cerevisiae Tma108 (scTma108) was compared to 14 well-characterized M1 aminopeptidases using multiple alignment of primary structures, followed by residue conservation analysis. The alignment shown corresponds to the most conserved region and is restricted to five M1 aminopeptidases. Residues in blue are fully conserved among the 15 M1 aminopeptidases (including Tma108), those in grey are partially conserved (90% of the sequences shown). Residues that are conserved in all the M1 aminopeptidases except in Tma108 are indicated in cyan on Tma108 sequence. Furthermore, Tma108 conservation was analyzed after retrieval of Tma108 orthologs in the diverse sequenced yeast genomes. Eighteen sequences from species (including S. cerevisiae) distributed along the phylogenic tree of the Saccharomycetaceae, were selected to detect residues that are fully conserved. These residues are indicated by color boxes at the top of the alignment (line labeled cons): dark blue boxes indicate residues that are conserved among Tma108 orthologs and similar in the reference aminopeptidases, orange boxes indicate residues that are specific to Tma108 orthologs. Numbers in the boxes indicate residues that were previously shown to be implicated in M1 aminopeptidase activities (based on the crystal structure of aminopeptidase N(42,43)): binding of the first amino acid of the substrate in S1 pocket (1), binding of the second amino acid of the substrate in S'1 pocket (2), binding of the Zn2+ cofactor (z), catalysis (c). Frames on the alignment indicate the two sequence signatures of the M1 family, (G/A/H/V)(G/A)MEN and HEXXHX18E, and the tyrosine involved in the catalytic mechanism. A dashed line indicates the separation between domains 1 and 2 of Tma108 inferred by homology based structure modeling (see below). (B and C) 3D molecular model of Tma108 of S. cerevisiae. Four domains are shown in different colors from light gray to black: NH2-terminus (1–227), central (228–490), hinge (491–578) and COOH terminal (579–946). Residues underscored by the two conservation analyses (b: conserved residues in all the M1 aminopeptidases, c: conserved residues in all the Tma108 orthologs) described in (a) are colored in the structure (same color code as in a).