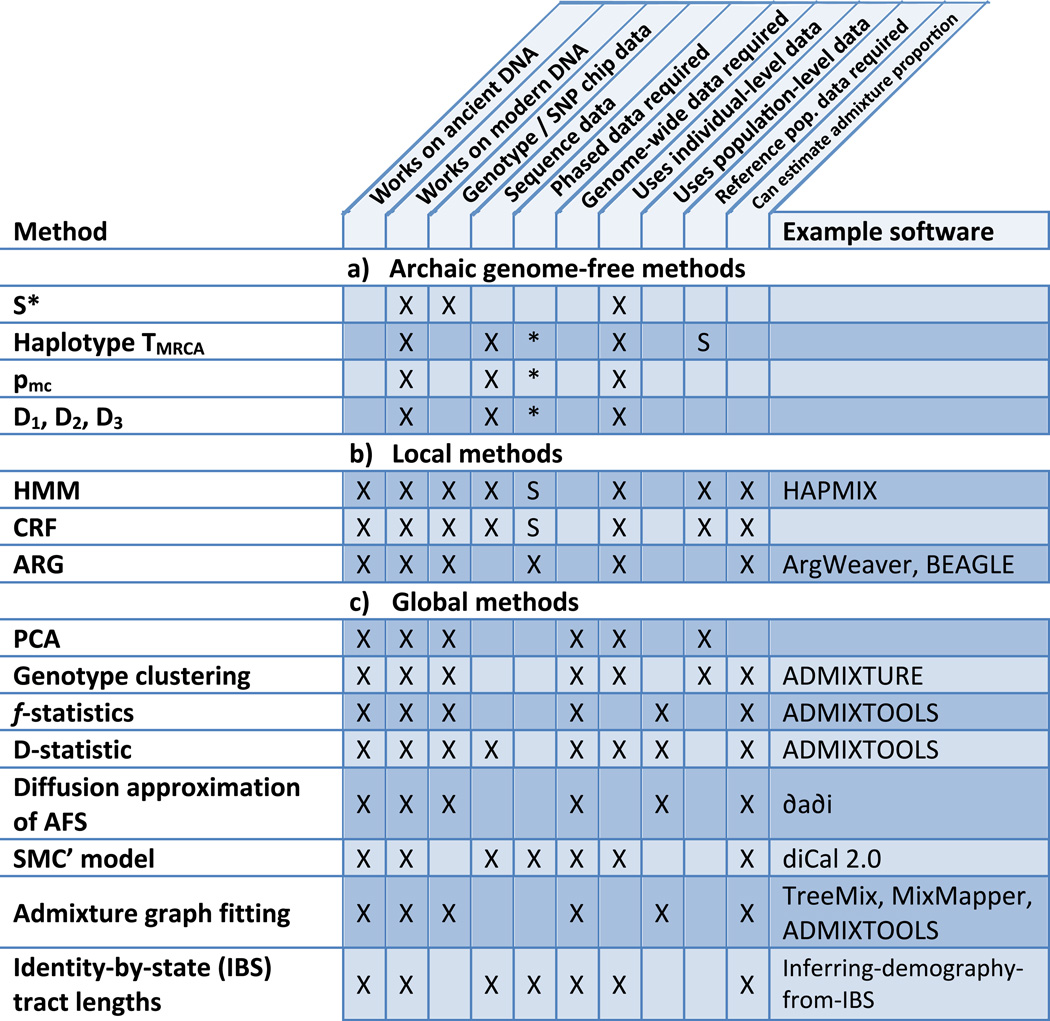
Figure 2.
Overview of popular techniques for studying archaic admixture. a: Archaic genome free methods are test statistics that can be used to infer archaic introgression into modern individuals without archaic sequence data. Each is computed on real data, then data simulated under various demographic models, and compared. These are prone to errors in model specification and can produce false positives. b: Local methods can be used to find specific genes or genomic regions admixed individuals derive from one or another ancestral population. These are tuned to detect detect long introgressed haplotypes but have reduced power to detect old admixture events. c: Global methods consider individual sites across the genome. Many are formal tests for admixture and/or can be used to estimate admixture proportion. In each box, “X” means true and “S” means true in some cases. “*” indicates methods applied to haplotype sequences, to which the concept of phasing does not apply. Note that, if sufficiently high-coverage genome-wide sequence data are available, these can be transformed into SNP calls if necessary. Also note that a method working on population-level data requires reference population data by default, as all inputs are population-level.