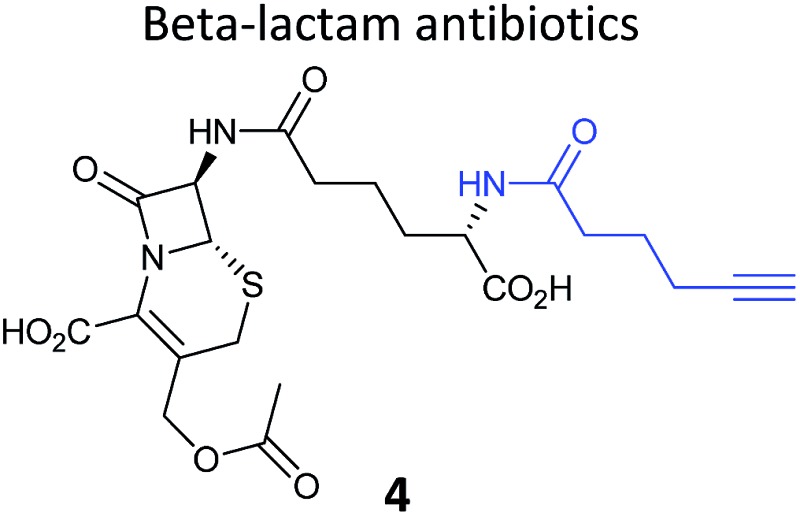
|
Pathogenic bacteria |
50
|
| Penicillin binding proteins and β-lactamase identified as targets |
| |
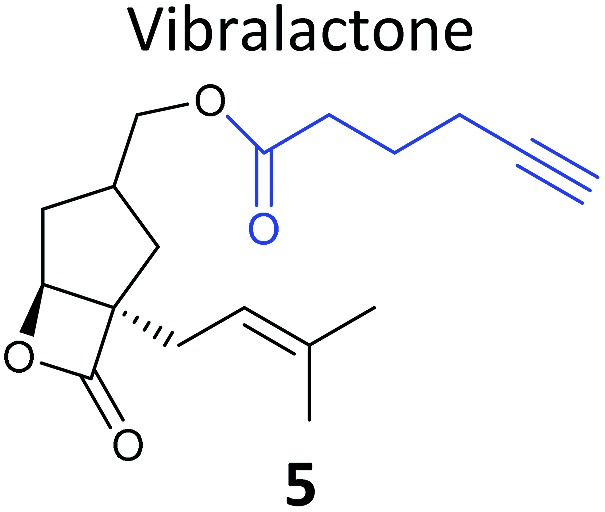
|
Listeria monocytogenes
|
56
|
| Multiple targets, including ClpP |
| |
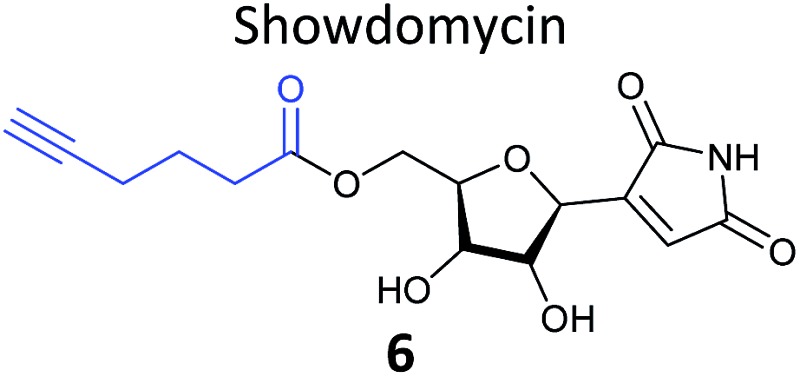
|
Pathogenic bacteria |
58
|
| Targets: oxidoreductases and transferases, including the enzyme MurA1 |
| |
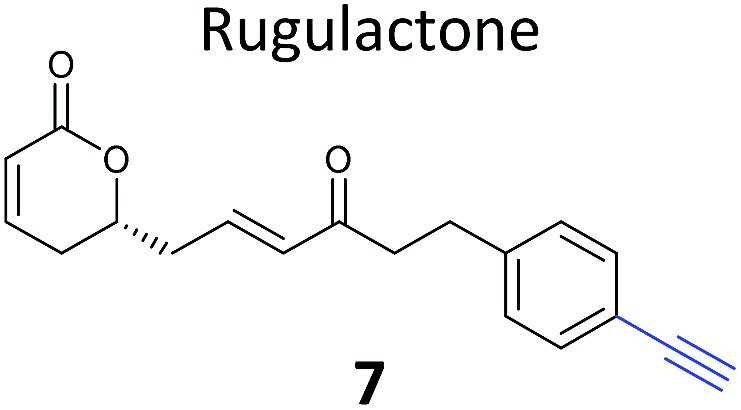
|
Pathogenic bacteria |
59
|
| Targets: multiple, including kinase ThiD |
| |
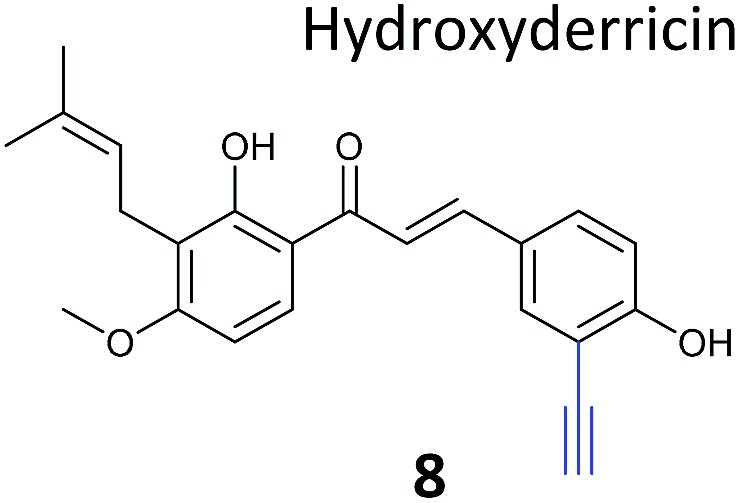
|
S. aureus
|
60
|
| Targets: multiple, including seryl-tRNA synthetase |
| |
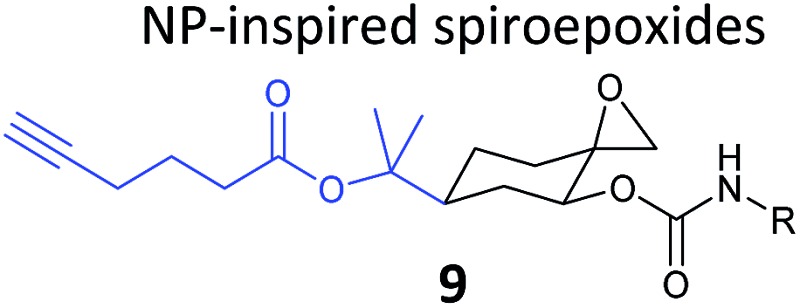
|
Human breast cancer cells |
62
|
| Target: phosphoglycerate mutase 1 (PGAM1) |
| |
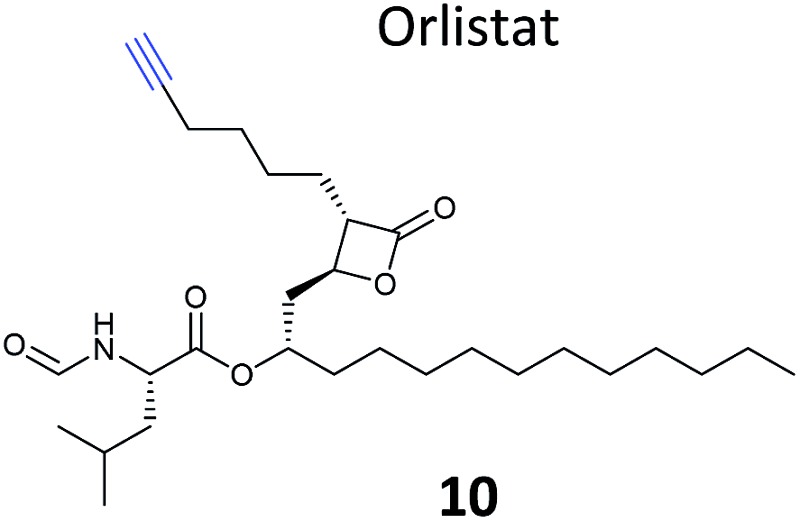
|
HepG2 liver cancer cells |
63
|
| Targets: multiple, including FAS (fatty acid synthase) and GAPDH |
| |
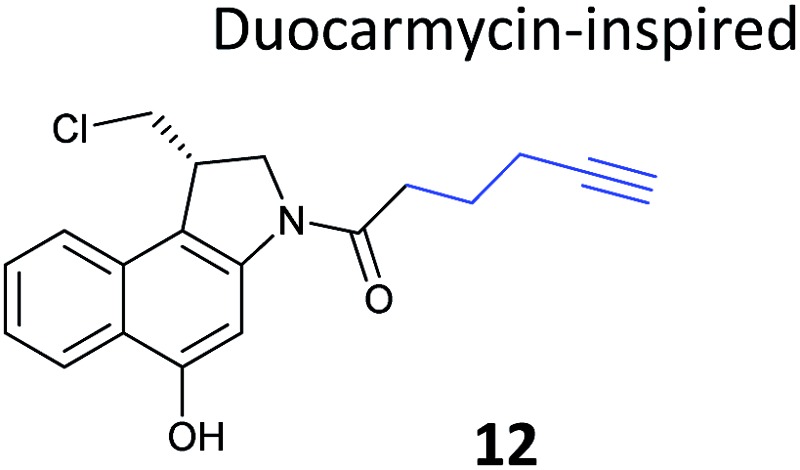
|
A549 cancer cells |
67
|
| Target: aldehyde dehydrogenase ALDH1A1 |
| |
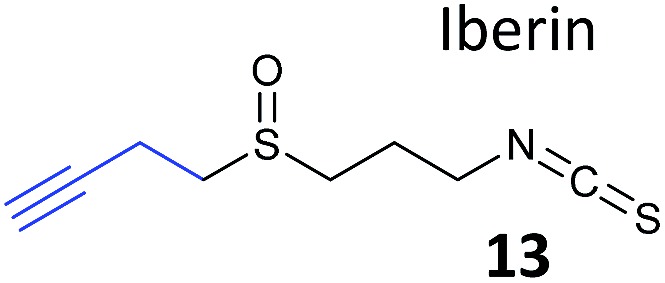
|
HEK293 cells expressing TLRs |
70
|
| No proteomics – immunoaffinity methods identified binding to TLRs |









