Fig. 5.
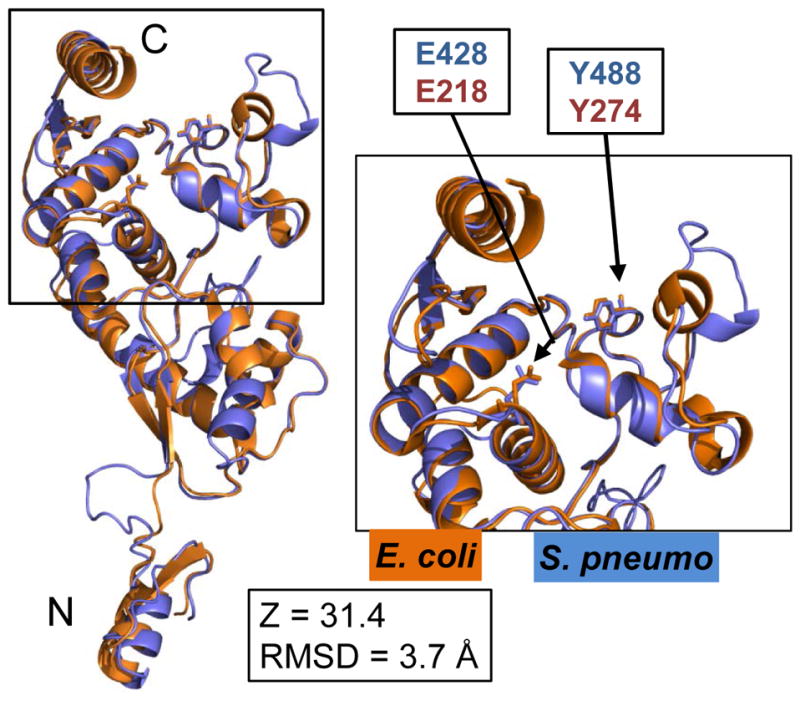
Superposition of the C-terminal YceG domains of MltGEco (orange, aa 81–340) and MltGSpn (blue, aa 266–547), including side chains of the conserved catalytic glutamate (E428 of MltGSpn; E218 of MltGEco) and tyrosine (Y488 of MltGSpn; Y274 of MltGEco). The MltGSpn structural model was generated by using the Phyre2 server as described in Experimental procedures, which listed aa 266–547 of MltGSpn alignment to aa 81–340 of MltGEco with 100% match confidence. The short N-terminal intracellular region and transmembrane of MltGEco (aa 1–80) were not included in the crystal structure analysis. The Phyre2-generated structural model of MltGSpn (blue) was overlaid and aligned with the crystal structure of the C-terminal of MltGEco (orange, PDB 2R1F) using PyMOL. Inset shows a close up of the fold with the conserved catalytic glutamate (E428/E218) and tyrosine (Y488/Y274). N and C indicate the amino- and carboxyl-ends of the aligned peptides. RMSD and Z-scores were obtained as described in Experimental procedures and Table S4.
