Figure 1.
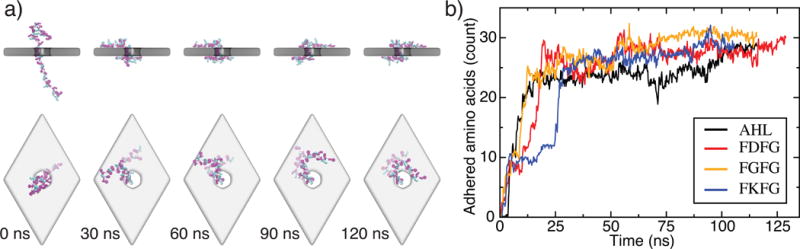
Equilibrium conformations of unfolded proteins threaded through a nanopore in a three-layer graphene membrane. (a) Equilibration simulation of the (FKFG)12 peptide. The sequence of snapshots illustrates the microscopic conformations of the peptide during the equilibration simulation. Graphene is shown as a gray transparent molecular surface The protein is shown using the licorice representation, with phenylalanine shown in magenta, lysine shown in teal, and glycine shown in white. At the beginning of the simulation, the protein is threaded through the nanopore and extended away from the membrane. The protein collapses onto the membrane in less than 30 ns and remains in contact with the membrane, diffusing along the membrane’s surface. (b) The number of amino acids adhered to the graphene membrane. Here, an amino acid is considered adhered to the membrane if its center of mass is located within 7 Å of the nearest carbon layer of the membrane, and it is not within the nanopore. Data are shown for a 48 residue fragment of α-hemolysin, negatively charged (FDFG)12, neutral (FGFG)12, and positive charged (FKFG)12 FG-Nups repeats. Each data point in this plot indicates a 200 ps block average of data sampled every 18-ps.
