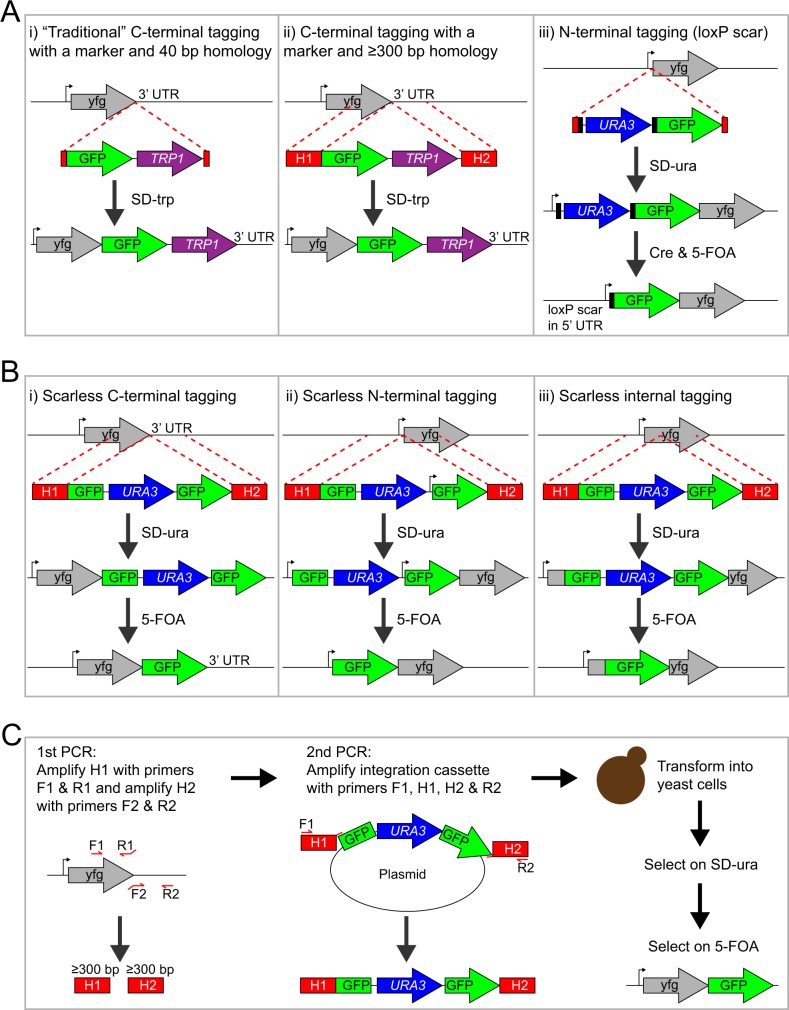
Fig 1. Cartoon of gene tagging methods in yeast.
(A) Commonly used methods for tagging a gene of interest (yfg = your favorite gene): C-terminal gene tagging using a marker and 40–50 bp homology (Fig 1A-i) or ≥300 bp homology (Fig 1A–ii), and N-terminal gene tagging using the Cre-loxP system (Fig 1A-iii). Methods i and ii use a selection marker that disrupts the 3’ UTR and cannot be eliminated, which might perturb the function of the fusion. For method iii, the loxP-flanked selection marker can be excised with the Cre recombinase, which leaves behind one flippase recognition target (FRT) site “scar” in the 5’ UTR of the tagged ORF. (B) Tagging methods introduced in this study: scarless C-terminal gene tagging (Fig 1B-i), scarless N-terminal gene tagging (Fig 1B-ii), and scarless internal gene tagging (Fig 1B-iii). The scarless tagging methods require a second round of selection to eliminate the URA3 marker, which is surrounded by identical GFP sequences. Note that the resulting “GFP scar” becomes a genuine part of the full-length GFP fusion protein after recombination and that the endogenous UTRs are not altered. The scarless N-terminal tagging method can be used for tagging essential genes in haploid yeast cells because a constitutive promoter situated between the URA3 marker and the second GFP fragment drives expression of the ‘partial GFP’-tagged gene prior to excision of URA3. Integration cassettes with either a partial GFP tag (as shown here) or a full-length GFP tag (S1 Fig) can be used. (C) Detailed description of the steps for scarless C-terminal tagging of a gene of interest with GFP. The integration cassette is built using two-step PCR synthesis. The homology arms H1 and H2 are amplified in the first round of PCRs and then used as primers together with F1 and R2 in the second round of PCRs. The primer-binding sites of H1 and H2 are unique, which is important for the efficient PCR amplification of the integration cassettes. Excision of the URA3 marker is not shown in this cartoon but is identical to the depiction above (Fig 1B-i).