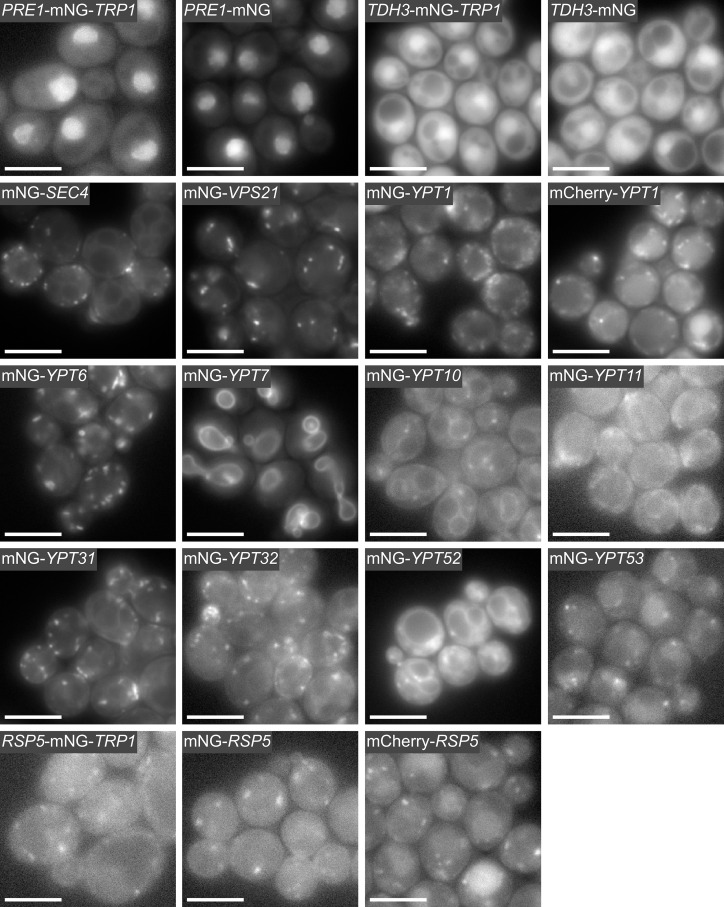
Fig 3. Fluorescence microscopy of S. cerevisiae cells producing mNeonGreen-tagged fusion proteins.
Yeast strains were constructed by modifying the endogenous gene locus using C-terminal gene tagging with a marker (Fig 1A-ii), scarless C-terminal gene tagging (Fig 1B-i), or scarless N-terminal gene tagging (Fig 1B-ii). PRE1 and TDH3 were tagged C-terminally with mNeonGreen (mNG) using the TRP1 selection marker or scarless. The eleven yeast Rab proteins (i.e. Sec4, Vps21, Ypt1, Ypt6, Ypt7, Ypt10, Ypt11, Ypt31, Ypt32, Ypt52, and Ypt53) are prenylated at their C-termini (S3 Fig) and were tagged N-terminally with mNeonGreen using the scarless gene tagging method. The Rab proteins localize to their expected organelles: for example Golgi (Ypt1, Ytp6), trans-Golgi network (Ypt6), vacuole and late endosomes (Ypt7), early endosomes (Vps21), recycling endosomes and post-Golgi exocytic vesicles (Ypt31), and secretory vesicles (Sec4). The ubiquitin ligase RSP5 was tagged C-terminally and scarless N-terminally with mNeonGreen. The C-terminal RSP5-mNeonGreen fusion resulted in cell size enlargement (S4 Fig) but no growth phenotype was observed (Fig 2I and S5 Fig). The large-cell phenotype was not observed with the scarless N-terminal fusions. The cells producing mCherry-Rsp5 and mCherry-Ypt1 fusions look similar to the cells producing the corresponding N-terminal mNeonGreen fusions, although the mCherry fusions also display some vacuolar localization. PRE1, YPT1, SEC4, and RSP5 are essential genes. Scale bar (white) is 5 μm.