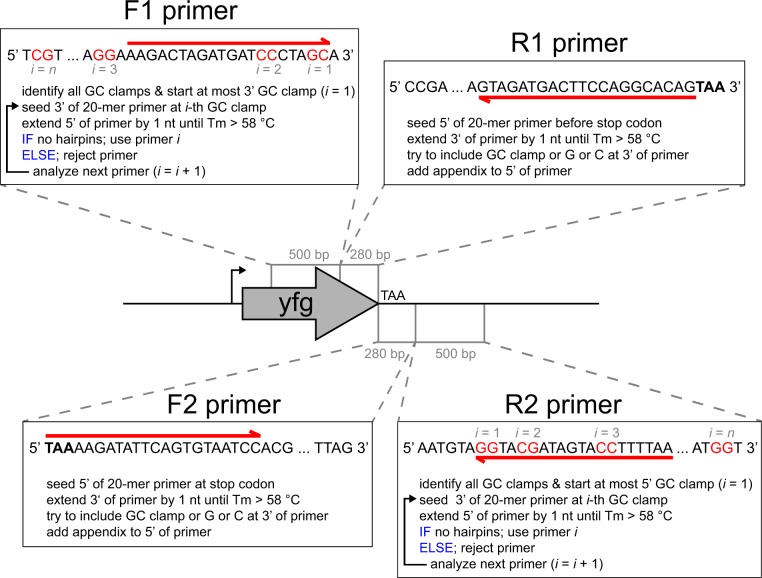
Fig 5. Pseudocode describing the automated primer design algorithm.
The illustration depicts the primer design for C-terminal gene tagging of a gene of interest (yfg) with a fluorescent protein. N-terminal and internal gene tagging is analogous except for the insertion site. The primers are shown as arrows (red) above and below the respective DNA sequences. GC clamps in the 500-bp upstream and downstream search regions are highlighted in red. The primer sequences for scarless N-terminal, scarless C-terminal, and C-terminal gene tagging with a marker for essentially all S. cerevisiae ORFs are provided in S4 Table. The primer sequences for scarless C-terminal tagging of all S. pombe ORFs are provided in S5 Table. The code for the automated primer design is contained in S1 File and available on GitHub (https://github.com/DXL38/scarless_gene_tagging_in_yeast). Primers F1 and R1 are used for amplifying the homology arm H1, and primers F2 and R2 are used for amplifying the homology arm H2. The homology arms H1 and H2 are ≥300 bp in size.