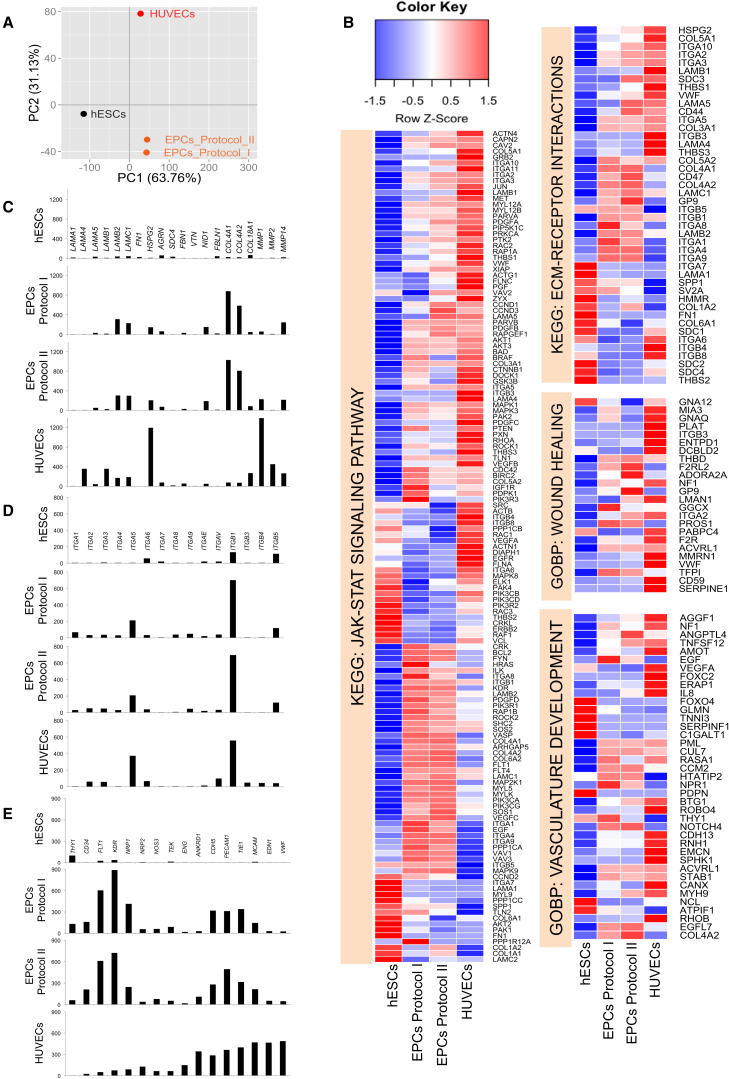
Figure 6.
Global Transcriptome Analyses of hESCs, hESC-Derived EPCs, and HUVECs
(A) Principal component analysis of 10,370 genes with FPKM >5 in at least one sample revealed 63.76% variability of the dataset along component 1 and 31.13% along component 2.
(B) Detailed gene-level heatmaps for key KEGG and GOBP biological pathways that are significantly upregulated in EPCs and HUVECs, compared with hESCs (FDR <5). Rows represent genes and columns represent samples. Row z-score transformation was performed on log2 of FPKM values with blue denoting a lower and red a higher expression level compared with the average expression level.
(C–E) Relative expression of extracellular matrix proteins (C), different α and β integrin subunits (D), and endothelial lineage markers (E) were plotted in their respective scales. y axis represents normalized FPKM values from RNA-seq.