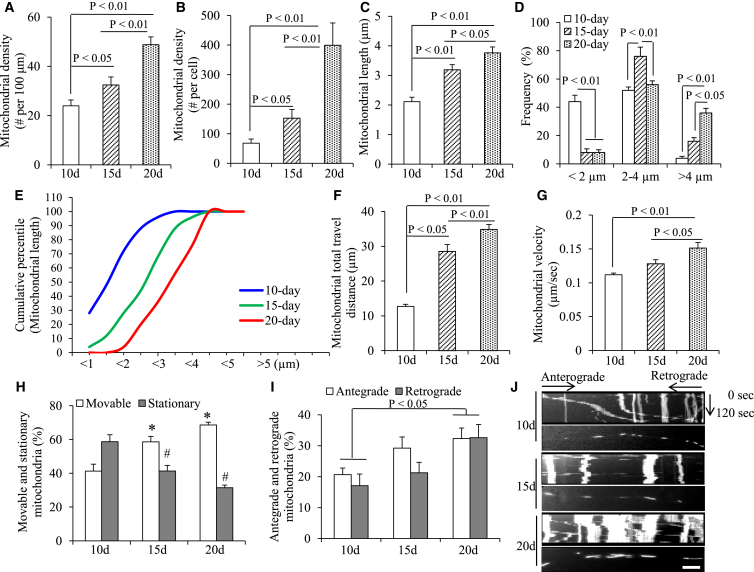
Figure 4.
Development of Mitochondrial Morphological and Dynamic Parameters in hiPSC-Derived DA Neurons
(A–D) Morphological changes in mitochondria of hiPSC-derived DA neurons cultured in differentiation media for 10, 15, and 20 days were measured and compared. Average mitochondrial density (in each 100-μm process in A and in all processes of each DA neuron in B), length (C), and frequency (D) in the processes of DA neurons were quantified.
(E) Cumulative distribution data showed increases in the numbers of long mitochondria and decreases in fragmentation of small mitochondria at 10, 15, and 20 days after differentiation induction (n = 3 independent experiments; mean ± SEM, with 10 mitochondria quantified per experiment in A–E).
(F–I) Average mitochondrial travel distance (F) and mitochondrial travel velocity (G) were calculated. The percentage of stationary (H), anterograde, and retrograde movable mitochondria (I) in DA neurons differentiated for varying days was compared. (n = 3 independent experiments; mean ± SEM, with 10 mitochondrial movements quantified per experiment in F–I).
(J) Kymographs generated from live imaging movies represent hiPSC-derived DA neurons cultured in differentiation media for 10, 15, and 20 days. In the kymographs, the x axis is mitochondrial position and the y axis represents the time lapse of 0–120 s. Vertical white lines represent stationary mitochondria and diagonal lines represent moving mitochondria. Anterograde movements are from left to right and retrograde movements are from right to left.
∗p < 0.05 versus day-10 movable mitochondria and #p < 0.05 versus day-10 stationary mitochondria in (H). Scale bar, 10 μm in (J).
Statistical analysis was performed using StatView version 5.0.1. For (A–D) and (F–I), one-way ANOVA was used for repeated-measures analysis, followed by Fisher's protected least significant difference for post hoc comparisons. Data are presented as mean ± SEM. Three independent experiments in (A–I); n = 10 per group per experiment in (A–I).