Figure 3.
Identification of RUNX1 and Genetic Editing of DS Cells
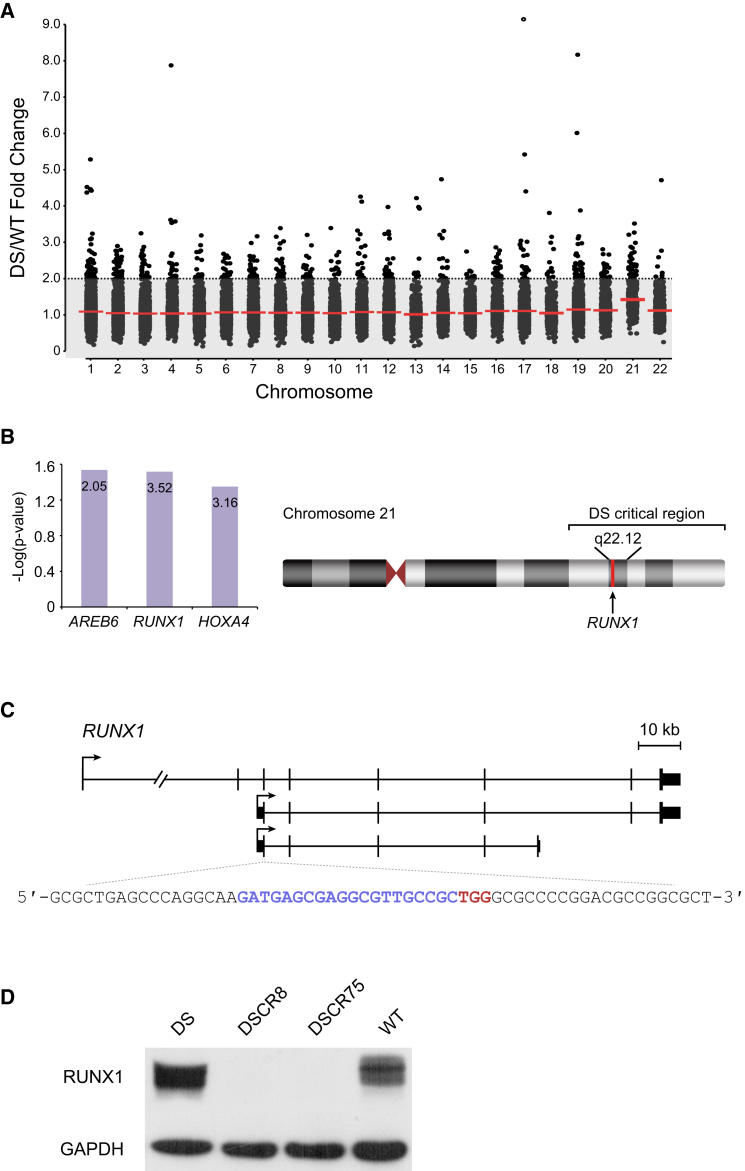
(A) Analysis of differentially expressed genes shows many of the upregulated genes of more than 2-fold expression to be located on different chromosomes. Each dot represents a gene and red lines represent average fold change in expression for each chromosome between DS- and WT-NPCs.
(B) RUNX1 was found to be a common transcription factor regulating many of the upregulated genes in DS-NPCs. Height of bars represents -log(p value) while the number inside the bar represents fold enrichment of the motif in the promoter set of the upregulated genes in DS-NPCs. RUNX1 resides at chromosome 21q22.12, within the critical region responsible for DS.
(C) Guide RNA was designed to target the exon common to all RUNX1 gene isoforms. Blue letters represent the guide sequence and red letters represent the PAM sequence of the guide RNA.
(D) Western blot analysis showing a complete ablation of RUNX1 in edited DS-ESCs (DSCR), clones DSCR8 and DSCR75 compared with their parental line (DS) and with WT-ESCs.