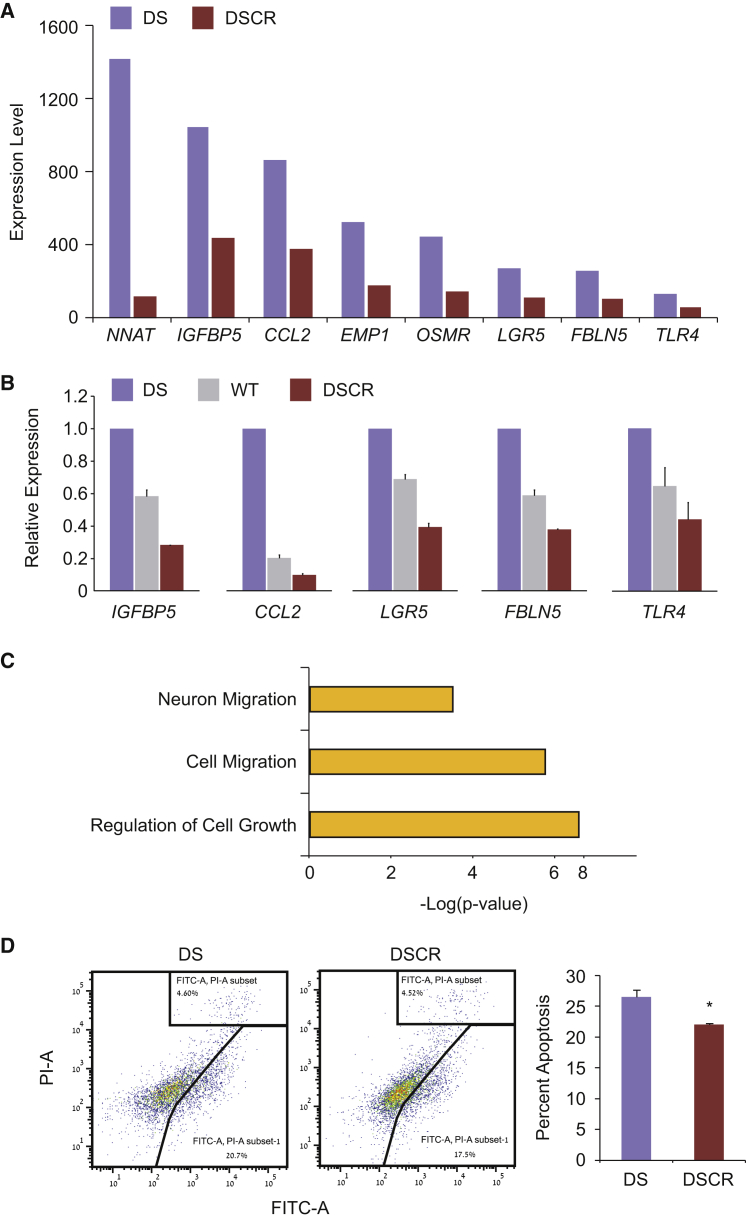
Figure 4.
Molecular and Cellular Analysis of Genomically Edited DS-NPCs
(A) DSCR-NPCs show downregulation of several key developmental genes, all putative targets of RUNX1. Microarray expression data of two DSCR cell lines were compared with microarray expression data of their isogeneic parental DS line. DSCR bars represent the average levels of expression of two DSCR microarrays.
(B) Several of these genes were further analyzed for their expression by qRT-PCR and showed a RUNX1 dosage-dependent expression pattern in DS-, WT-, and DSCR-NPCs. For DS, the parental cell line was used in three independent experiments, for DSCR, the two isogenic cell lines were used in three independent experiments, and for WT, three different cell lines were used. Error bars represent SEM.
(C) Functional annotation analysis of all downregulated genes in DSCR-NPCs shows significant enrichment for neuron migration, cell migration, and regulation of cell growth.
(D) Comparison of the apoptotic levels of the isogenic DS- and DSCR-NPCs were analyzed and show downregulation of apoptosis in the edited cells as seen by the summation of annexin V+(FITC+)/PI− and annexin V+(FITC+)/PI+ cell populations in DSCR-NPCs (middle panel, representing one cell line) and DS-NPCs (left panel, representing one cell line). Bar graph represents the average summation of annexin V+/PI− and annexin V+/PI+ populations as percentage of apoptotic cells. For DS the parental cell line was used in three independent experiments, and for DSCR the two isogenic cell lines were used in three independent experiments. Error bars represent SEM. ∗p < 0.05 using Student's t test.