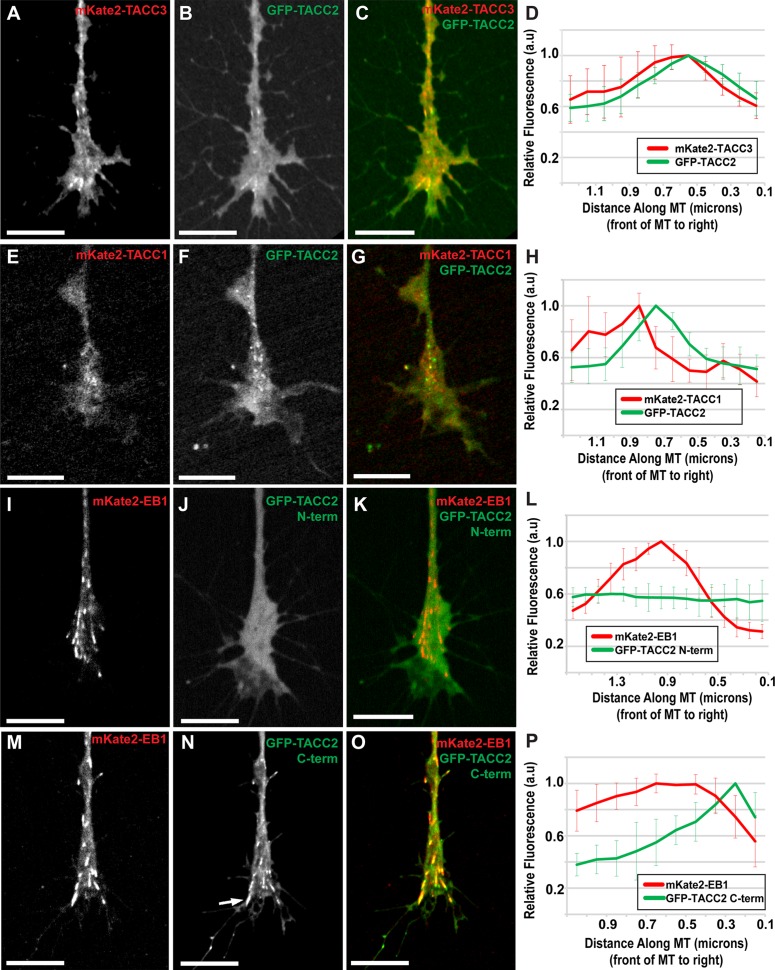
FIGURE 2:
TACC2 localizes to the plus end through its C-terminal domain and overlaps with both TACC1 and TACC3. (A–C) Representative images of mKate2-TACC3, GFP-TACC2, and merged in cultured neuronal growth cones. (D) Fluorescence intensity profiles of GFP-TACC2 and mKate2-TACC3. (E–G) mKate2-TACC1, GFP-TACC2, and merge with GFP-TACC2 fluorescence intensity plot profile (H). (I–K) Images of mKate2-EB1, GFP-TACC2 N-term (aa 1–540), and merged with the plot of fluorescence intensity profiles (L). (M–O) Images of mKate2-EB1, GFP-TACC2 C-term (aa 923-1168), and merged with the plot showing their fluorescence intensity profiles (P). Arrow in N points to lattice binding of TACC protein. Time interval between all frames is ∼2 s. For D, the green plot profile was translated ∼0.2 μm left (MT plus-end velocity ∼0.124 μm/s, green channel imaged 1.5 s after red) to accurately represent relative localization. For H, the green profile was translated left based on the velocity of each comet measured to account for difference in acquisition time points between channels (green channel imaged 1.5 s after red). In P, the green profile was translated 0.1 μm (MT plus-end velocity ∼0.148 μm/s, red channel imaged 0.96 s after green). For all fluorescence intensity profiles, ∼10 MTs for each condition were quantified by intensity line scans, and SDs are shown in error bars. MT plus end is at the right. Scale bars, 10 μm.