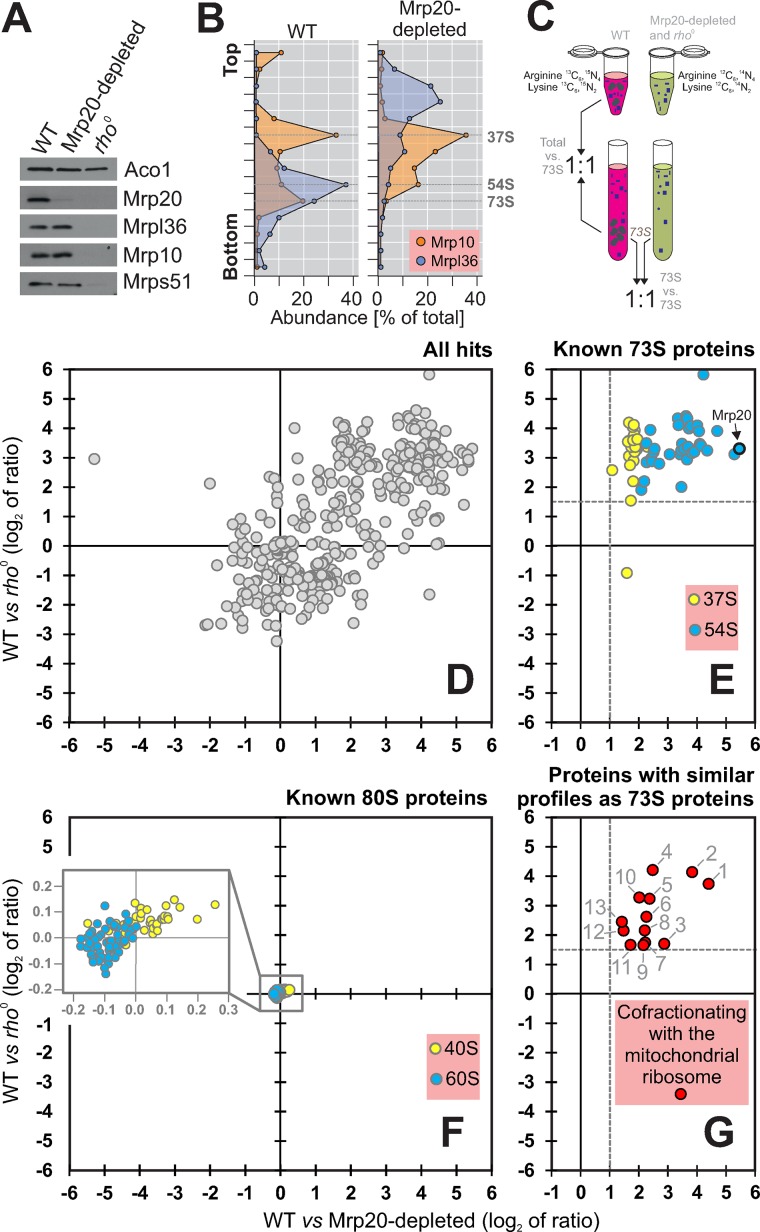
FIGURE 1:
Identification of proteins cofractionating with mitochondrial ribosomes. (A) Proteins of the large (Mrp20, Mrpl36) and small (Mrp10, Mrps51) ribosomal subunits, as well as of aconitase (Aco1), were analyzed by Western blotting of mitochondrial extracts isolated from the indicated strains. (B) Mitochondria were isolated from wild-type and Mrp20-depleted cells, lysed, and separated on a linear sucrose gradient. Proteins of 16 fractions were analyzed by Western blotting with Mrp10- and Mrpl36-specific antibodies. The signals were quantified. The graph shows their distribution in the gradient. (C) Schematic overview of the differential profiling strategy. (D–G) For the SILAC analysis, wild-type cells were grown in the presence of “heavy” (13C6, 15N4-arginine, 13C6, 15N2-lysine) medium. The rho0 cells and cells of the Mrp20-depleted mutant were grown in “light” (12C6, 14N4-arginine, 12C6, 14N2-lysine) medium. Mitochondrial extracts were separated on a linear sucrose gradient. The 73S-containing fractions were pooled and their protein profiles analyzed by mass spectrometry. (D) Plot of the log2 ratios of all 466 identified proteins obtained in these two experiments. (E, F) Proteins of the large (blue) and small (yellow) subunits of mitochondrial (E) and cytosolic (F) ribosomes. (G) Proteins that cofractionated with mitochondrial ribosomes. The proteins shown were all enriched at least threefold in the 73S fraction over the total extract of wild-type mitochondria.