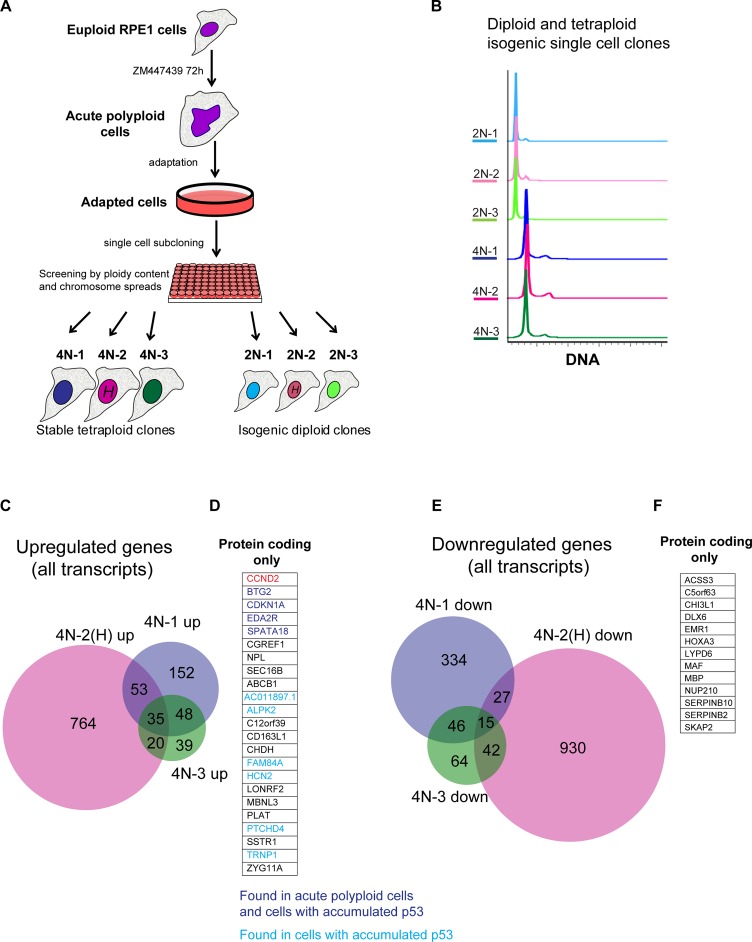
FIGURE 4:
Generation of proliferating tetraploid single-cell clones from acute polyploid Aurora-inhibited cells and analysis of their transcriptome by RNAseq. (A) Generation of proliferating tetraploid RPE1 single cell clones (4N 1–3) from euploid RPE1 cell culture treated with Aurora inhibitor ZM447439. Asynchronously growing cells were treated with ZM447439 for 72 h and allowed to survive for several weeks, until proliferating populations emerged. Proliferating cells were plated in 96-well plates at a density of one cell per well. The ploidy content of each single-cell clone was analyzed by FACS. Isogenic diploid single-cell clones (2N 1–3) were picked from the same 96-well plates as the corresponding tetraploid clones. Single-cell clones 4N-2 and 2N-2 underwent Hoechst 33342 labeling (H) before subcloning. (B) Ploidy analysis of generated tetraploid and isogenic diploid single-cell clones. Cells were fixed and stained with propidium iodide. FACS profiles indicate doubling of the DNA content. (C) Venn diagram of genes up-regulated in stable tetraploid single-cell clones in comparison to isogenic diploid single-cell clones. Only genes with log2 FC ≥ 1, p adjusted ≤ 0.001, and expression values log2 average RPKM > 0 are included. (D) Expanded list of genes from C, showing names of overlapping genes up-regulated in all three clones (protein-coding transcripts only). Genes that were previously found to be up-regulated in acute polyploid cells (Aurora inhibited and anillin knockdown) are highlighted in dark blue. Genes that were previously found to be up-regulated only in p53-accumulated cells treated with nutlin-3 are highlighted in light blue. (E) Venn diagram of genes down-regulated in stable tetraploid single-cell clones in comparison to isogenic diploid single-cell clones. Includes genes with log2 FC ≤ −1, p adjusted ≤ 0.001, log2 average RPKM > 0. (F) Expanded list of overlapping genes found in E, showing names of genes down-regulated in all three tetraploid single-cell clones (protein-coding transcripts only).