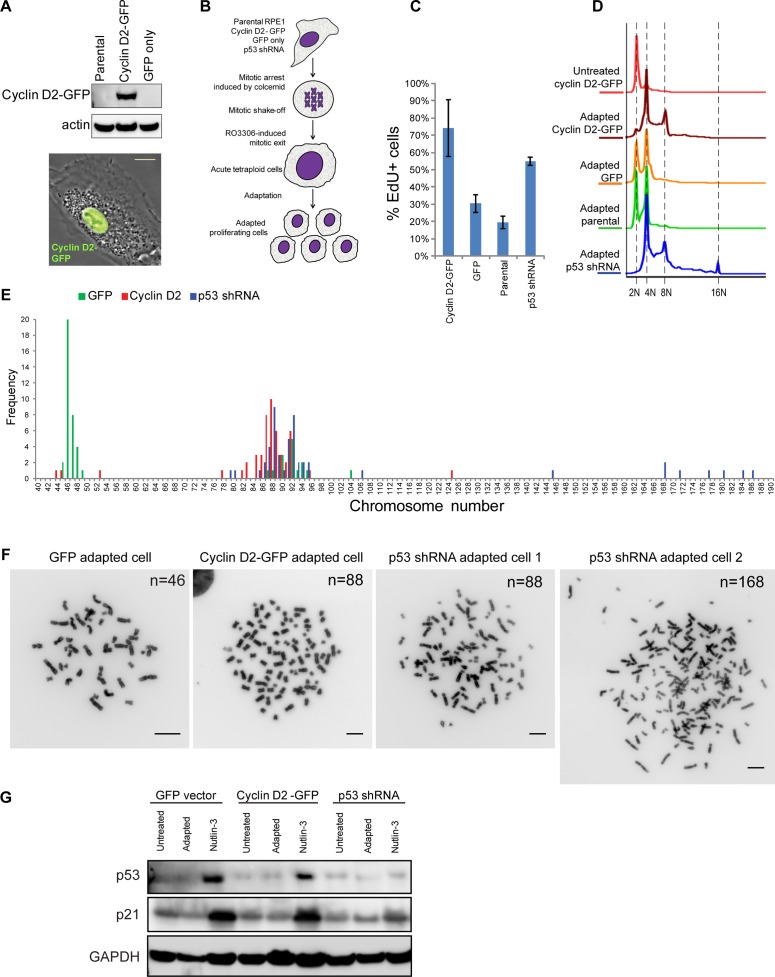
FIGURE 8:
Experimental adaptation to tetraploidy after forced mitotic exit. (A) Western blot (top) and phase contrast/fluorescence image (bottom) of RPE1 cells overexpressing cyclin D2–GFP. Bar, 10 μm. (B) Design of experimental adaptation to tetraploidy after forced mitotic exit. Adaptation experiments included cells overexpressing cyclin D2-GFP, GFP empty vector, p53 shRNA, and parental RPE1 cell lines of early passage. Mitotic cells were collected after incubation in Colcemid for 6 h and treated with 10 μM CDK1 inhibitor RO-3306 to induce mitotic exit. After drug washout, cells were left to survive until proliferating populations emerged. Each experiment was performed in triplicate. (C) EdU incorporation assay of adapted cells overexpressing cyclin D2-GFP, cells expressing GFP empty vector, parental cells, and cells expressing p53 shRNA. Cells were allowed to incorporate EdU for 24 h. Bar heights represent the mean (n = 4); error bars denote SD. (D) Ploidy distributions of adapted cells overexpressing cyclin D2-GFP, cells expressing GFP empty vector, parental cells, and cells expressing p53 shRNA. Cells were fixed and stained with propidium iodide. Histograms indicate doubling of the DNA content predominantly in cells overexpressing cyclin D2-GFP and cells expressing p53 shRNA. (E) Chromosome counts from adapted populations of cells overexpressing cyclin D2–GFP or GFP alone and expressing p53 shRNA. Fifty spreads were counted from each group. (F) Representative mitotic chromosome spreads from every adapted population; n, chromosome number. Bar, 10 μm. (G) Western blot analysis of p53 and p21 protein levels in cells expressing GFP, cyclin D2–GFP, and p53 shRNA before adaptation (untreated), after adaptation, and treated with 2.5 μM nutlin-3 for 24 h.