MenaINV, an actin-regulatory protein known to promote metastasis, has roles in driving cross-talk between extracellular matrix, growth factor cues, and their downstream pathways during tumor cell invasion. MenaINV is a shared component of the signaling pathway driving both EGF chemotaxis and fibronectin haptotaxis.
Abstract
Directed cell migration, a key process in metastasis, arises from the combined influence of multiple processes, including chemotaxis—the directional movement of cells to soluble cues—and haptotaxis—the migration of cells on gradients of substrate-bound factors. However, it is unclear how chemotactic and haptotactic pathways integrate with each other to drive overall cell behavior. MenaINV has been implicated in metastasis by driving chemotaxis via dysregulation of phosphatase PTP1B and more recently in haptotaxis via interaction with integrin α5β1. Here we find that MenaINV-driven haptotaxis on fibronectin (FN) gradients requires intact signaling between α5β1 integrin and the epidermal growth factor receptor (EGFR), which is influenced by PTP1B. Furthermore, we show that MenaINV-driven haptotaxis and ECM reorganization both require the Rab-coupling protein RCP, which mediates α5β1 and EGFR recycling. Finally, MenaINV promotes synergistic migratory response to combined EGF and FN in vitro and in vivo, leading to hyperinvasive phenotypes. Together our data demonstrate that MenaINV is a shared component of multiple prometastatic pathways that amplifies their combined effects, promoting synergistic cross-talk between RTKs and integrins.
INTRODUCTION
Directed cell motility is required for many physiological processes and can be driven by various cues. One example is the process of metastasis—the dissemination of cells from a primary tumor to secondary sites in the body—which is responsible for 90% of deaths associated with cancer. Directional cues induce the local invasion of tumor cells into adjacent tissue and vasculature, ultimately leading to metastatic dissemination. Chemotaxis—the directional movement of cells attracted to a source of soluble cues that diffuse passively onto the cells—has been studied extensively in many systems (Bear and Haugh, 2014). In particular, epidermal growth factor (EGF) secreted by tumor-associated macrophages acts as a powerful chemotactic attractant for nearby breast tumor cells (Wyckoff et al., 2004). Extracellular matrix (ECM) proteins also drive directed migration by haptotaxis, by which cells move in response to differences in the concentration of substrate-bound signals. The ECM protein fibronectin (FN) is abundant in tumor tissue, particularly around the vasculature and at the tumor periphery (Astrof et al., 2004; Zhou et al., 2008), and guides haptotaxis in breast cancer and melanoma (Chan et al., 2014; Oudin et al., 2016b). The plethora of directional cues within the tumor microenvironment raises the question of how cells integrate their migration responses to these signals to metastasize efficiently.
Cells generally encounter multiple soluble and substrate-bound cues simultaneously in vivo. It is well established that the ECM binds growth factors (GFs), forming a GF reservoir that contributes to the generation of stable GF gradients (Hynes, 2009). Furthermore, ECM proteins themselves can activate receptor tyrosine kinases (RTKs), which occurs either via binding of intrinsic ECM domains such as EGF-like repeats to EGF receptor (EGFR) directly or via RTK transactivation by integrins (Kuwada and Li, 2000; Balanis and Carlin, 2012). Finally, integrins and RTKs have been shown to signal together in the context of tumor cell invasion. The mutant version of the tumor suppressor p53, which is mutated in 50% of cancers, drives invasion through enhanced Rab coupling protein (RCP)–dependent recycling of α5β1, the main receptor for FN, and several RTKs, EGFR, and Met (Caswell et al., 2008; Muller et al., 2013). Although the potential for cross-talk between ECM and GF cues has been demonstrated, its role in haptotaxis and the relationship between chemotaxis and haptotaxis remains poorly understood.
The actin-regulatory protein Mena, a member of the Ena/VASP family of proteins, regulates cell migration and has been implicated in both chemotaxis and haptotaxis (Bear and Gertler, 2009; Gertler and Condeelis, 2011). Mena is up-regulated in breast cancer, and deletion of Mena in the PyMT-MMTV genetic model of breast cancer reduces metastasis (Roussos et al., 2010). During tumor progression, Mena is alternatively spliced to produce multiple functionally distinct isoforms. MenaINV, which contains a 19–amino acid inclusion, is heterogeneously expressed in breast tumors and is associated with increased metastasis and poor clinical outcome, especially compared with expression of Mena alone (Oudin et al., 2016a, b). Mechanistically, MenaINV increases sensitivity to various GFs present within the tumor microenvironment, including EGF, hepatocyte growth factor (HGF), and insulin growth factor (IGF), by dysregulating the recruitment of the tyrosine phosphatase PTP1B to cognate receptors after ligand stimulation. Consequently expression of MenaINV causes increased receptor phosphorylation and corresponding cell invasion in response to low levels of ligand stimulation (Hughes et al., 2015). Second, Mena isoforms, and especially MenaINV, drive metastasis through their ability to support haptotaxis on FN gradients via direct α5 integrin interaction (Gupton et al., 2012; Oudin et al., 2016b). In particular, MenaINV allows cells to haptotax at high FN concentrations similar to those present around blood vessels and in the tumor periphery via increased outside-in signaling at focal complexes (FXs) and inside-out ECM remodeling (Oudin et al., 2016b). Given its role in regulating responses to GFs and FN, we hypothesized that MenaINV could promote α5β1-EGFR cross-talk via receptor transactivation, recycling, or synergy. Here we demonstrate that MenaINV is a shared component of the machinery that mediates both haptotactic responses to FN and chemotactic responses to EGF.
RESULTS
Mena- and MenaINV-driven haptotaxis requires EGFR signaling
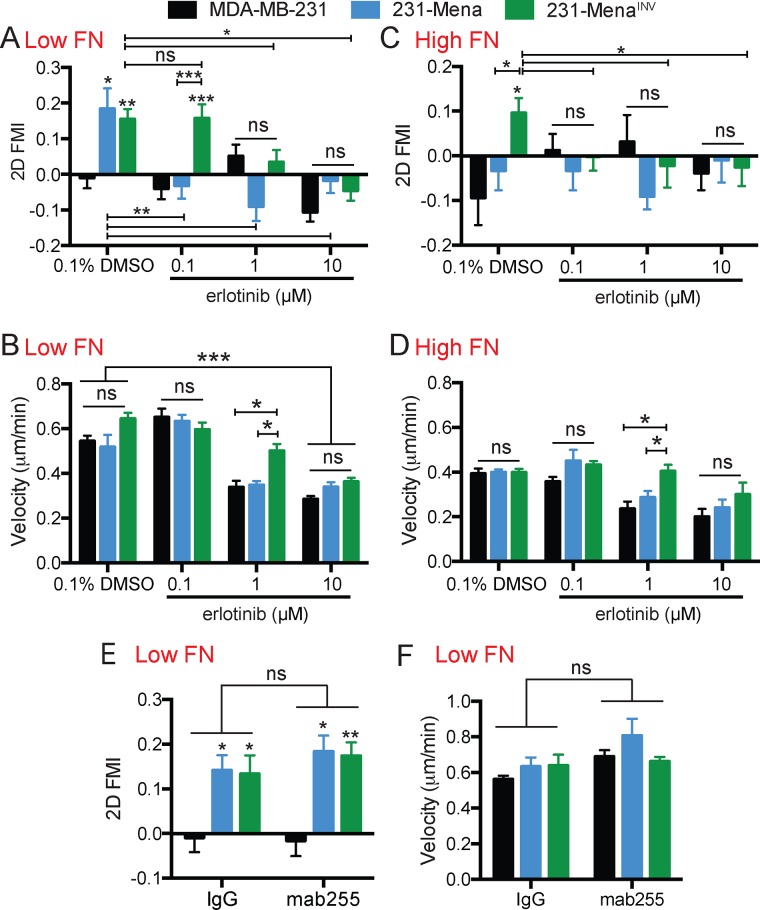
Given the established role of MenaINV in driving GF sensitization (Hughes et al., 2015) and promoting haptotaxis on FN gradients (Oudin et al., 2016b) and the emerging role of RTKs in response to ECM cues (Zhu and Clark, 2014), we asked whether MenaINV-driven haptotaxis on FN gradients requires EGFR signaling. Cultured breast tumor cells express low levels of Mena and undetectable levels of MenaINV relative to the amounts found in invasive cells collected from these tumors (Goswami et al., 2009; Oudin et al., 2016a). We generated cell lines from the p53-mutant, triple-negative breast cancer cell line MDA-MB-231, which stably express Mena or MenaINV at levels similar to the amount of Mena found in invasive tumor cells (Wang et al., 2007; ∼10-fold overexpression with respect to endogenous Mena; cells referred to as 231-Control, 231-Mena, or 231-MenaINV). Previously we used a microfluidic device (Wu et al., 2012) to study cells migrating on FN gradients by time-lapse imaging, quantify the forward migration index (FMI), and assess their directional movement relative to the FN gradient (details in Supplemental Figure S1A). Given our recent findings that MenaINV enhances chemotactic response to EGF by dysregulating EGFR signaling, we wondered whether the EGFR signaling pathway might be involved in haptotaxis. Consistent with this possibility, erlotinib, an EGFR-specific kinase inhibitor, blocked Mena-driven haptotaxis on low FN gradients (125 μg/ml FN at the top of the gradient; Figure 1, A and B), whereas 231-MenaINV cells required a 10-fold higher concentration of erlotinib than 231-Mena cells to block haptotaxis completely (Figure 1A). Of interest, the effects on haptotaxis were distinct from those on cell velocity: 0.1 μM erlotinib blocked Mena-driven haptotaxis without affecting velocity, whereas higher erlotinib concentrations affected both haptotaxis and cell velocity (Figure 1B). We confirmed these findings using a second EGFR kinase inhibitor, gefitinib (Supplemental Figure S1B). To confirm the specificity of the inhibitors, we used small interfering RNA (siRNA) to transiently knock down EGFR in the cell lines (Supplemental Figure S1C). Knockdown of EGFR inhibited Mena- and MenaINV-driven haptotaxis while significantly decreasing cell velocity (Supplemental Figure S1, D and E). We previously found evidence that MenaINV expression enables haptotaxis at high FN concentrations (500 μg/ml FN at the top of the gradient; Oudin et al., 2016b). Here we found that erlotinib also inhibited MenaINV haptotaxis at high FN concentrations (Figure 1C). The overall velocity of all cells was reduced on high FN gradients, but again the effects on velocity versus directionality were distinct: 0.1 μM erlotinib blocked MenaINV-driven haptotaxis without affecting velocity (Figure 1D). We then investigated whether the haptotaxis-promoting EGFR activity on FN gradients required ligand-mediated receptor activation. Addition of mab225, which blocks EGF-dependent activation of EGFR but not its intrinsic kinase activity (Meyer et al., 2013), had no effect on haptotaxis or velocity (Figure 1, E and F), suggesting that the requirement for EGFR kinase activity revealed by sensitivity to erlotinib was not dependent on activation of EGFR by ligand. Together these data suggest that MenaINV-driven haptotaxis on low and high FN gradients requires EGFR signaling but not ligand-elicited EGFR activity.
FIGURE 1:
Mena/MenaINV-driven FN haptotaxis requires EGFR. (A) Inhibition of EGFR with different concentrations of erlotinib decreased Mena- and MenaINV-driven haptotaxis on a 2D 125 μg/ml low FN gradient, as measured by the FMI. (B) Velocity (μm/min) of 231, 231-Mena, and 231-MenaINV cells treated with increasing concentrations of erlotinib. (C) Inhibition of EGFR with different concentrations of erlotinib decreased MenaINV-driven haptotaxis on a 2D high 500 μg/ml FN gradient. (D) Velocity (μm/min) of 231, 231-Mena, and 231-MenaINV cells treated with increasing concentrations of erlotinib on a high FN gradient. (E) FMI and (F) velocity of MDA-MB-231, 231-Mena, and 231-MenaINV cells migrating on 2D low FN gradient with an immunoglobulin G control of mAB225. Data pooled from at least 50 cells from three different experiments. Results show mean ± SEM; significance by one-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.005. Asterisks above a column in the graph characterize significance relative to control within that condition. See Supplemental Figure S1.
MenaINV-driven haptokinesis requires EGFR signaling
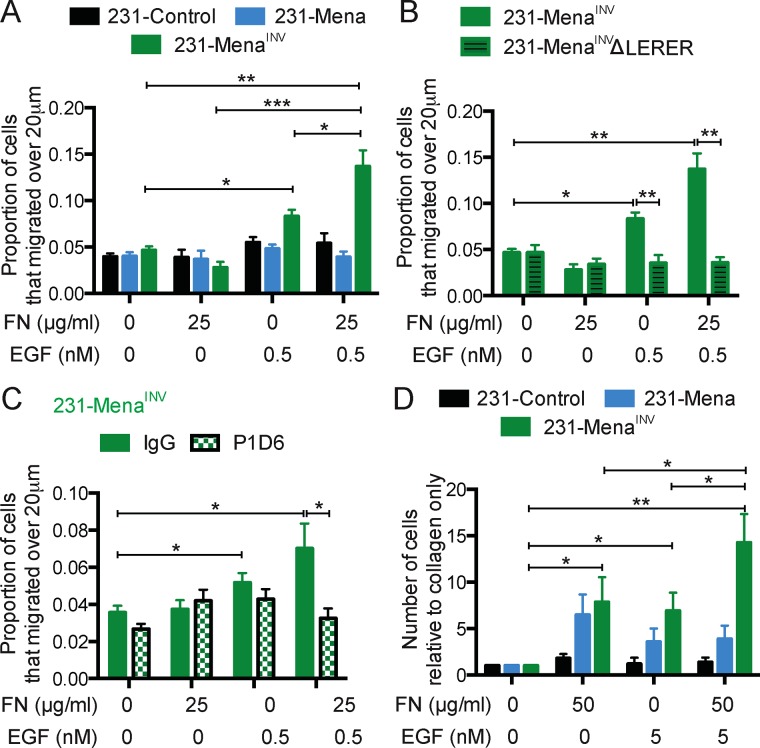
Having shown that EGFR signaling is required for MenaINV-driven two-dimensional (2D) haptotaxis on FN gradients, we investigated whether this pathway was also important in haptokinesis—random migration induced by ECM cues. It is well known that FN can induce migration of cells; however, it does so in a biphasic manner, with highest levels of migration at intermediate concentrations (DiMilla et al., 1993). To investigate whether MenaINV plays a role in FN haptokinesis, we examined cellular invasion in three-dimensional (3D) collagen gels containing uniform FN at different concentrations (Supplemental Figure S2A). We found that 231-Control and 231-Mena cells exhibited a biphasic response to FN concentration, with maximum invasion elicited by 50 μg/ml FN and migration returning to baseline at 125 μg/ml (Supplemental Figure S2, B and C). In contrast, 231-MenaINV cells still invaded at maximal levels at the higher 125 μg/ml FN concentration, indicating that 231-MenaINV cells have a shifted dose–response relation for FN (Figure 2A; see Supplemental Figure S2F for vehicle control). We then investigated the role of the interaction between MenaINV and the cytoplasmic tail of α5 integrin in haptokinesis. The LERER region is unique to Mena and mediates direct binding to the unique C-terminal portion of the cytoplasmic tail of α5 and not other α integrin subunits; deletion of LERER abrogates the interaction with α5 and has distinct functional consequences for adhesion signaling and migration (Gupton et al., 2012; Oudin et al., 2016b). Similar to our previous observations for MenaINV-driven haptotaxis, we found that direct Mena–α5 integrin interaction was required for MenaINV-driven haptokinesis, as 231-MenaINV ΔLERER cells did not display increased invasion in response to any of the FN concentrations tested (Supplemental Figure S2D). The apparent lack of invasion in the 50 μg/ml condition by 231-MenaINV ΔLERER cells suggests that this construct may have dominant-negative effects that block the modest proinvasive activity, for example, by sequestering ligands bound to its other domains in the absence of the ability to interact with α5. Inhibition of α5β1 with blocking antibody P1D6 inhibited FN-driven invasion of 231-MenaINV cells (Supplemental Figure S2E). In contrast, Cilengitide-mediated inhibition of αvβ3, another major FN receptor, had no effect (Supplemental Figure S2E). We then asked whether EGFR signaling was involved in MenaINV-driven invasion into FN and found that erlotinib inhibited invasion of 231-MenaINV cells at all FN concentrations (Figure 2B). Together these data suggest that MenaINV-driven FN haptokinesis requires both α5β1 function and EGFR kinase activity.
FIGURE 2:
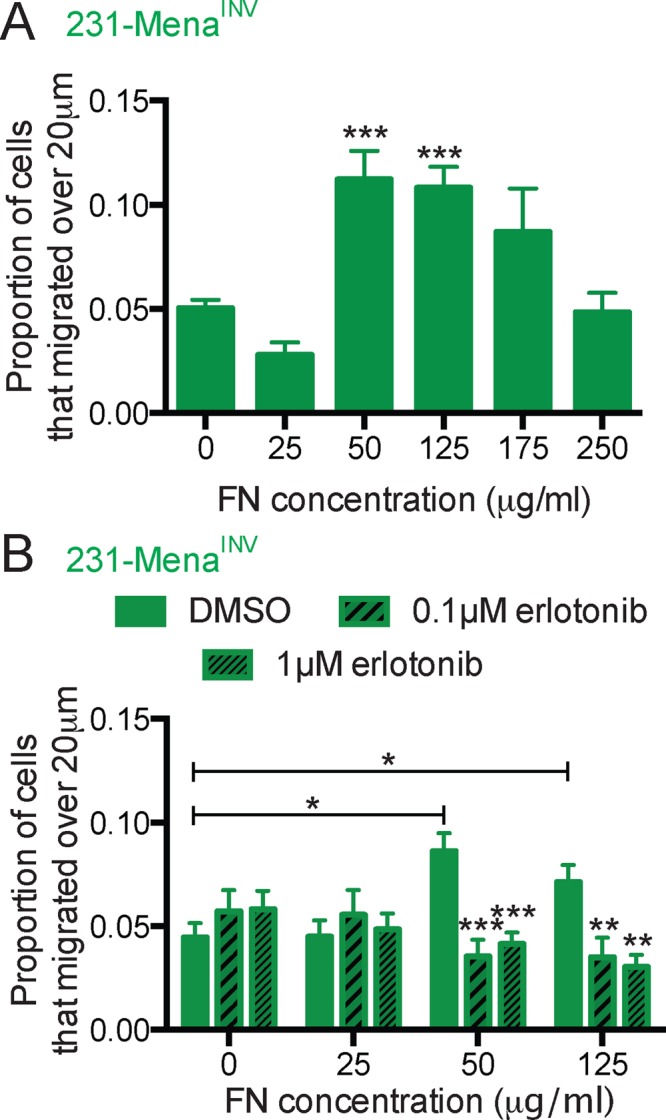
EGFR signaling is important for MenaINV-driven FN haptokinesis. (A) In vitro invasion assay in a 3D collagen gel with increasing concentration of FN (0, 25, 50, 125, 175, 250 μg/ml) for 231-MenaINV cells treated with serum-free medium for 24 h. The proportion of cells migrating 20 μm above baseline was quantified. (B) Treatment with erlotinib (0.1 or 1 μM) decreased the GF-independent effect of FN on 3D invasion in 231-MenaINV cells. Mean ± SEM, results pooled from at least five different experiments; significance determined by one-way ANOVA, * p < 0.05, ** p < 0.01, *** p < 0.005. See Supplemental Figure S2.
MenaINV expression promotes EGFR signaling on FN
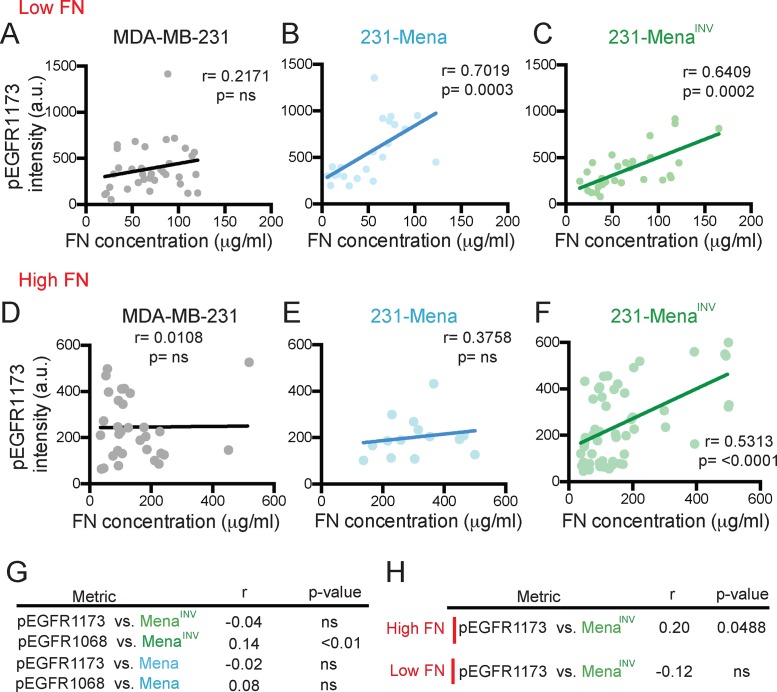
The requirement for EGFR signaling during haptotaxis prompted us to ask whether Mena and MenaINV drove EGFR phosphorylation in cells migrating on FN gradients. In cells haptotaxing on FN gradients, we quantified levels of EGFR phosphorylation at Y1173, a residue exhibiting increased phosphorylation in 231-MenaINV after EGF stimulation (Hughes et al., 2015). First, we confirmed that expression of Mena, MenaINV, and variants with a deletion of the LERER domain did not affect expression of EGFR (Supplemental Figure S3, A–C). Using immunofluorescence, we quantified pEGFRY1173 levels in cells plated on gradients of FN of different concentrations. On a low FN gradient, FN concentration correlated positively and significantly with pEGFRY1173 in 231-Mena and 231-MenaINV cells but not in control MDA-MB-231 cells (Figure 3, A–C). This correlation was absent when cells were treated with erlotinib (Supplemental Figure S3, D–F). However, at high FN concentration, only 231-MenaINV cells showed a significant correlation between pEGFRY1173 and FN (Figure 3, D–F). We then investigated whether Mena and MenaINV mRNA expression in human breast cancer patients was associated with phosphorylation of EGFR, using The Cancer Genome Atlas (TCGA) database (Cancer Genome Atlas Network, 2012). There was a significant correlation between levels of MenaINV, but not Mena, and levels of pY1068, but not pY1173, in human breast cancer patients from TCGA with available reverse-phase protein array data (Figure 3G). Of interest, in patients with high tumor FN mRNA levels, there was a weak but statistically significant correlation between MenaINV and pEGFRY1173 (Figure 3H). Together these data suggest that high FN and MenaINV levels are correlated with increased EGFR activity.
FIGURE 3:
MenaINV drives EGFR signaling on FN. (A–C) Correlation between pEGFR1173 in whole cell relative to cell area and FN concentration under the cell in cells plated on a low 125 μg/ml FN gradient for MDA-MB-231 (A), 231-Mena (B), and 231-MenaINV cells (C). (D–F) Correlation between pEGFR1173 in whole cell relative to cell area and FN concentration under the cell in cells plated on a low 125 μg/ml FN gradient for MDA-MB-231 (D), 231-Mena (E), and 231-MenaINV cells (F). Data from at least 15 cells per cell line pooled from three different experiments. (G) Correlations (Spearman’s r and p value) between Mena or MenaINV and pEGFR1173 or pEGFR1068 from the reverse-phase protein assay (RPPA) data in the TCGA breast cancer cohort. (H) Correlations (Spearman’s r and p value) between Mena or MenaINV and pEGFR1173 or pEGFR1068 from the RPPA data in the TCGA breast cancer cohort in patients with high or low FN. See Supplemental Figure S3.
Inhibition of PTP1B phenocopies MenaINV expression and drives haptotaxis
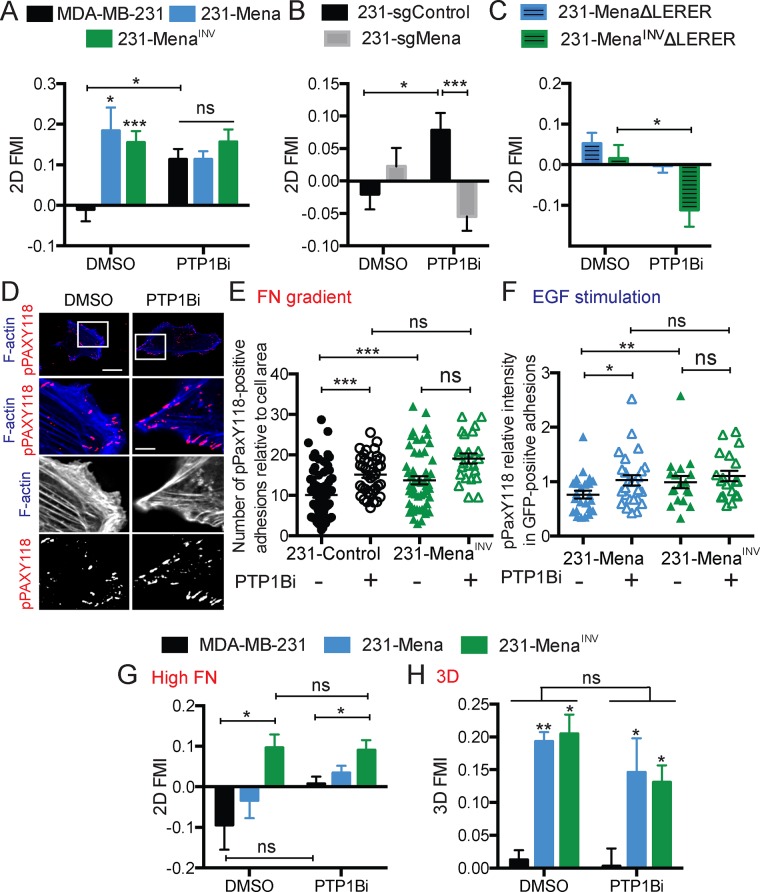
MenaINV expression affects sensitivity to proinvasion growth factors by dysregulating the phosphatase PTP1B; pharmacological inhibition of PTP1B (PTP1Bi) in parental MDA-MB-231 cells phenocopies many EGF-related MenaINV-driven phenotypes (Hughes et al., 2015). We therefore asked whether PTP1Bi could also drive haptotaxis of parental MDA-MB-231 in different contexts. PTP1Bi induced haptotaxis of MDA-MB-231 cells on a low 2D FN gradient without affecting the migratory response of 231-Mena or 231-MenaINV cells (Figure 4A). We examined whether the PTPBi effects on signaling alone would support haptotaxis in the complete absence of even the low levels of endogenous Mena in parental MDA-MB-231 cells. We found that clustered regularly interspaced short palindromic repeats–mediated deletion of Mena (231-sgMena cells) indeed blocked this effect (Figure 4B; knockout validation in Supplemental Figure S4A), suggesting that the prohaptotatic influence of PTP1Bi required endogenous Mena expression. Furthermore, the interaction between Mena/MenaINV and α5 was also necessary for this prohaptotactic effect because 231-MenaΔLERER and 231- MenaINVΔLERER cells did not haptotax in the presence of PTP1Bi (Figure 4C).
FIGURE 4:
Inhibition of PTP1B phenocopies MenaINV. (A) FMI of MDA-MB-231, 231-Mena, and 231-MenaINV cells treated with PT1Bi (10 μM) on a low 125 μg/ml FN gradient. (B) FMI of MDA-MB-231 cells with sgRNA targeted against a control sequence or Mena treated with PT1Bi (10 μM) on a low 125 μg/ml FN gradient. (C) FMI of 231-Mena ΔLERER and 231-MenaINVΔLERER cells treated with PT1Bi (10 μM) on a low 125 μg/ml FN gradient. (D) Representative images of 231-Control cells showing GFP (green), F-actin (blue), and pPAX118 (red) for cells plated on a low 125 μg/ml FN gradient and treated with PT1Bi (10 μM). Scale bar, 5 μm (main image), 1 μm (inset). (E) Number of pPaxY118-positive adhesions in 231-Control and 231-MenaINV cells plated on a 2D FN gradient with DMSO or PTP1Bi treatment. (F) Levels of pPaxY118 in Mena- positive adhesions in 231-Mena and 231-MenaINV cells plated on collagen and Matrigel and treated with 0.25 nM EGF for 1 min. (G) FMI of MDA-MB-231, 231-Mena, and 231-MenaINV cells treated with PT1Bi (10 μM) on a high 500 μg/ml FN gradient. (H) FMI of MDA-MB-231, 231-Mena, and 231-MenaINV cells treated with PT1Bi (10 μM) in a 3D 125 μg/ml FN gradient. Haptotaxis data are pooled from at least three experiments, with at least 75 cells analyzed overall. Results show mean ± SEM; significance determined by one-way ANOVA, * p < 0.05, ** p < 0.01, *** 0.005. See Supplemental Figure S4.
Because PTP1B is known to influence signaling at focal adhesions (FXs; Burdisso et al., 2013), we hypothesized that the haptotactic effect from PTP1Bi was associated with altered behavior of the key FX signaling regulator paxillin. We therefore performed immunofluorescence studies on haptotaxing cells plated on a 2D FN gradient for 3 h. We measured the number of pPAXY118-positive adhesions in cells expressing different Mena isoforms plated on a 2D FN gradient. 231-MenaINV cells, which haptotax toward high FN concentrations, had a significantly increased number of pPAX-positive adhesions relative to 231-Control cells (Figure 4, D and E). To test whether the haptotaxis effect of PTP1Bi is due to changes in phosphorylation of proteins in adhesions, we measured the number of pPAXY118-positive adhesions in 231-Control treated with PTP1Bi and found an increase with drug treatment (Figure 4, D and E).
Given that PTP1Bi increases EGFR phosphorylation downstream of EGF stimulation (Hughes et al., 2015), we next investigated whether MenaINV could also promote FX signaling downstream of RTK activation. After EGF stimulation, 231-MenaINV cells show increased FX-associated pPAXY118 compared with 231-Mena (Figure 4F). In addition, PTP1Bi significantly increased pPAXY118 in 231-Mena to similar levels as seen in 231-MenaINV (Figure 4F). Overall these data suggest that MenaINV drives PTP1B dysregulation and leads to increased FX signaling downstream of both chemotactic and haptotactic pathways.
Compared to Mena, MenaINV causes more sustained FX signaling and enables haptotaxis at high FN concentrations (Oudin et al., 2016b). Therefore we asked whether PTP1Bi could drive haptotaxis of MDA-MB-231 and 231-Mena cells on high FN gradients and found that PTP1Bi had no effect in this context (Figure 4G). This suggests that the increase in FX signaling is not sufficient to support haptotaxis on high FN concentrations. Furthermore, we found that PTP1Bi failed to drive haptotaxis of MDA-MB-231 cells in three dimensions (Figure 4H). Taken together, these data confirm that MenaINV can regulate FX signaling downstream of multiple cues and suggest that there are additional functional requirements for 3D versus 2D haptotaxis.
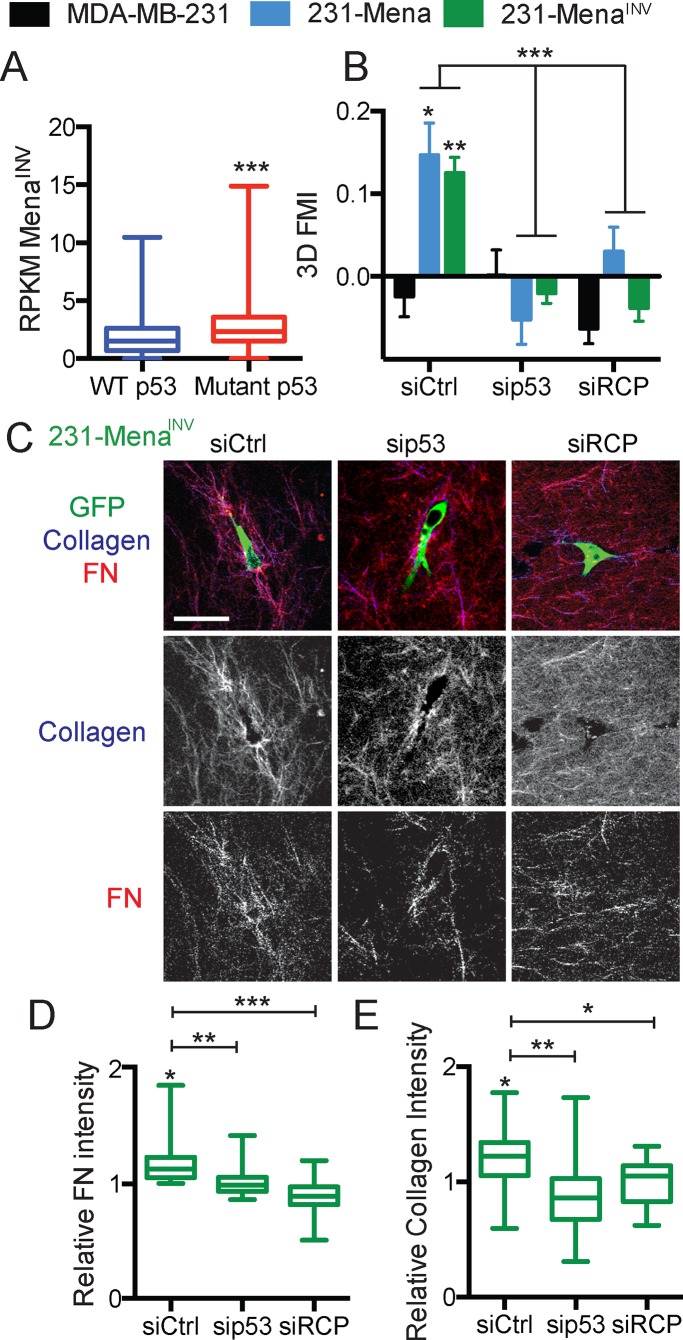
Knockdown of p53 and RCP, known to drive cotrafficking and recycling of α5β1 and EGFR, inhibits MenaINV-driven haptotaxis and ECM reorganization
Whereas converging on signaling at adhesions is one way in which EGFR/α5β1 may contribute to motility, another way in which EGFR/α5β1 cross-talk could contribute to 3D invasion is via their dysregulated cotrafficking (Muller et al., 2011). Recycling of cotrafficked α5β1/EGFR complexes is induced by expression of mutant p53 expression, which triggers RCP-mediated association between the two receptors, leading to increased invasion (Muller et al., 2009). Cells with mutant p53, such as the MDA-MB-231 cells used here, show increased levels of α5β1/EGFR recycling and preferential targeting of the recycled complex to the pseudopods of invading cells in three dimensions. Of interest, by examining data from the TCGA breast cancer cohort, we found that breast cancer patients with mutant p53 showed significantly higher levels of MenaINV than those with wild-type p53 (Figure 5A). Therefore we investigated whether dysregulated RCP-dependent recycling of α5β1/EGFR might also be involved in 3D haptotaxis by asking whether expression of either mutant p53 or RCP was required for MenaINV-driven 3D haptotaxis (Muller et al., 2009). First, we confirmed that there was no difference in level of total RCP or p53 expression in cells expressing Mena or MenaINV (or their corresponding LERER deletion mutants; Supplemental Figure S4, B–E). Knockdown of either p53 or RCP (Supplemental Figure S4, F–H), which was previously shown to block 3D invasion and recycling of α5β1/EGFR (Muller et al., 2009), decreased MenaINV-driven haptotaxis in three dimensions (Figure 5B). Three-dimensional haptotaxis also requires ECM reorganization (Oudin et al., 2016b), reflected by increased accumulation of FN and collagen around haptotaxis-competent cells in 3D gels. Knockdown of p53 and RCP led to a significant decrease in accumulation of FN and collagen around 231-MenaINV cells (Figure 5, C–E). Taken together, these data show a requirement for RCP and p53 in MenaINV-driven 3D haptotaxis and ECM reorganization, consistent with a model in which cotrafficking of α5β1/EGFR complexes supports these phenotypes.
FIGURE 5:
Knockdown of RCP and p53 inhibits MenaINV-driven haptotaxis and ECM reorganization. (A) Reads per kilobase of transcript per million mapped reads for MenaINV in TCGA breast cancer patients with WT or mutant p53. (B) Knockdown of p53 and RCP (72 h) inhibits Mena- and MenaINV-driven 3D FN haptotaxis. (C) Representative images of 231-MenaINV cells (GFP) in a 3D 125 μg/ml FN gradient (red) in a collagen gel (blue) with siCtrl, sip53, or siRCP. Scale bar, 25 μm. Knockdown of p53 and RCP (72 h) inhibited 231-MenaINV–driven increased accumulation and reorganization of (D) FN and (E) collagen. Results show mean ± SEM; significance by one-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.005. See Supplemental Figure S4.
MenaINV promotes synergy between EGF and FN
Our data suggest that MenaINV promotes directional migration downstream of α5β1 and RTKs. However, given that both cues are present simultaneously within the tumor microenvironment (Joyce and Pollard, 2009), we wondered how cells would respond to exposure to both cues simultaneously. We first used an in vitro chemoinvasion assay (described in Supplemental Figure S1); in this instance, collagen plus FN gels were overlaid with varying EGF concentrations. We verified the presence of gradients using fluorescent dextran of similar molecular weight to EGF (Supplemental Figure S5). Addition of a low-dose EGF gradient (0.5 nM) in the presence of 25 μg/ml FN/collagen did not affect the invasion of 231-Control or 231-Mena cells; however, these conditions evoked a 3D invasion response by 231-MenaINV cells that was significantly greater than that observed for addition of either 0.5 nM or 25 μg/ml FN individually (p = 0.039, two-way analysis of variance [ANOVA] interaction term; Figure 6A). These data suggest that MenaINV promotes synergy between FN and EGF. We then investigated the role of the Mena–α5 interaction in this synergistic response. Of interest, deletion of the LERER region in Mena or MenaINV did not affect protrusion of lamellipodia in response to low-dose EGF stimulation of cells plated on collagen and Matrigel, suggesting that acute actin polymerization in response to growth factor stimulation is intact in the absence of interaction with α5 (Supplemental Figure S6). However, in three dimensions in the presence of FN, the synergistic effect was eliminated by either abrogating the interaction between MenaINV and α5β1 (in 231-MenaINVΔLERER cells; Figure 6B) or inhibiting α5β1 with the P1D6 function blocking antibody (Figure 6C). To test this effect in vivo, we used an in vivo invasion assay, which allows for collection of cells from primary tumors by chemotaxis and invasion into microneedles filled with growth factors and various ECM factors (Wyckoff et al., 2000). We generated xenograft tumors by injecting MDA-MB-231 cells expressing Mena isoforms in the mammary fat pad of immune-compromised mice. The in vivo invasion assay revealed that the combination of EGF and FN attracted more cells from 231-MenaINV tumors than did each cue alone (Figure 6D). At the same concentrations, the combination of EGF and FN did not have a synergistic effect on invasion in 231-Control or 231-Mena tumors. Overall these data suggest that MenaINV promotes invasion by sensitizing cells to both soluble chemotactic signals and substrate-bound ECM signals and that MenaINV, via its α5 -interaction, enables a superadditive combined response.
FIGURE 6:
MenaINV promotes synergy between FN and EGF. (A) In vitro chemotaxis assay in a 3D collagen gel with EGF (0.5 nM), FN (25 μg/ml), or both for 231-Control, 231-Mena, and 231-MenaINV cells. The proportion of cells migrating 20 μm above baseline is quantified. (B) 3D chemotaxis of 231-MenaINV and 231-MenaINVΔLERER cells with FN, EGF, or both. (C) Treatment with the α5-blocking antibody P1D6 blocked EGF/FN synergy in a 3D invasion assay. (D) In vivo invasion assay in tumors generated in NOD/SCID mice with MDA-MB-231 cells expressing Control-GFP, Mena, or MenaINV. Needles contain 1 mg/ml collagen and increasing amounts of FN or EGF (four mice per condition). Results show mean ± SEM; significance determined by one-way ANOVA, * p < 0.05, ** p < 0.01, *** 0.005. See Supplemental Figures S5 and S6.
DISCUSSION
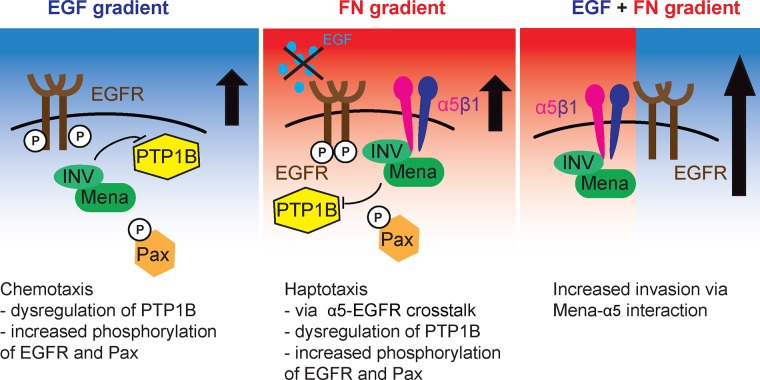
Here we investigated EGFR and α5β1 signaling cross-talk in the context of EGF chemotaxis and FN haptotaxis. We found that the actin-binding protein MenaINV participates in ECM/GF cross-talk in three ways: FN-driven EGFR transactivation, synergistic activation downstream of EGFR and α5β1, and recycling of EGFR and α5β1. Taken together, these data suggest that MenaINV is a shared component of the EGF chemotaxis and FN haptotaxis responses, two distinct prometastatic pathways driving local invasion in the tumor microenvironment.
First, our data point toward a role for MenaINV in promoting integrin and RTK signaling cross-talk downstream of both α5β1 and EGFR via receptor transactivation. MenaINV-driven 2D haptotaxis on FN requires EGFR activity driven by dysregulated phosphatase PTP1B. Correspondingly, PTP1Bi drives 2D haptotaxis on FN gradients in a Mena-dependent manner. These data, combined with prior observation that MenaINV dysregulates PTP1B-dependent attenuation of EGFR signaling, fit a model in which PTP1B facilitates α5β1/EGFR cross-talk observed during 2D haptotaxis. PTP1B is known to be present at FXs and can be found in complexes of β1 and β3 integrin (Burdisso et al., 2013). Furthermore, PTP1B is required for integrin-dependent down-regulation of RhoA and up-regulation of Rac1 (Burdisso et al., 2013). Therefore our data are consistent with PTP1B playing roles downstream of both RTKs and integrins. ECM/GF cross-talk, such as ligand-independent activation of RTK signaling, has been described in multiple contexts. For instance, FN-mediated activation of α5β1 leads to HGF-independent activation of Met to promote invasion (Mitra et al., 2011). Furthermore, α5β1-dependent phosphorylation of platelet-derived growth factor-β (PDGFR-β) is induced upon cell adhesion to FN in the absence of any growth factor (Veevers-Lowe et al., 2011). Given that Met, PDGFR, and IGF1R are substrates for PTP1B and that MenaINV increases sensitivity to HGF and IGF, it is possible that MenaINV-driven ECM/GF talk may extend to other RTKs. The data presented here point to MenaINV as a novel mediator of ECM-driven RTK transactivation via PTP1B dysregulation.
Second, we find that MenaINV promotes synergistic responses to combined EGF and FN gradients. Of interest, phospho–mass spectrometry analysis in 231-MenaINV cells treated with EGF revealed increased phosphorylation of multiple proteins known to be present at FXs and playing a role in responses to ECM cues, such as vinculin, paxillin, P130Cas, and FAK (focal adhesion kinase) (Hughes et al., 2015). Inhibition of PTP1B enables haptotaxis and increases FX signaling downstream of both EGF and FN. It is well established that signaling downstream of RTKs and integrins can converge on common signaling pathways, such as the Ras–mitogen-activated protein kinase pathway, the phosphoinositide 3-kinase/Akt pathway, or Rho GTPases, particularly at focal adhesions (Huttenlocher and Horwitz, 2011). However, how these pathways are coordinated to drive directional migration in response to multiple cues is poorly understood. Recent studies identified several important components of haptotactic responses that are actually not required for GF chemotaxis, including LKB1 signaling via kinases of the microtubule affinity-regulating kinase family (Chan et al., 2014). Of interest, two key cytoskeletal molecules—the Arp2/3 complex, which nucleates branched F-actin, and fascin, which supports filopodia formation through its F-actin bundling activity—are also similarly required for haptotaxis on FN gradients (Wu et al., 2012; Johnson et al., 2015) but not for chemotaxis to growth factors. Therefore, whereas some signaling pathway components are exclusively required downstream of a single directional cue, others, like MenaINV, may play roles in multiple modes of migration.
Finally, one other well-described mechanism by which RTK/integrin cross-talk can promote invasion is through cotrafficking (Paul et al., 2015). Mutant p53 has been shown to drive p53-dependent recycling of α5β1 with EGFR and Met, a process that contributes to 3D invasion (Muller et al., 2013). We previously reported that 231-MenaINV cells show a 30% increase in surface α5 levels (Oudin et al., 2016b) and a small but significant 10% increase in EGFR recycling (Hughes et al., 2015). Knockdown of p53 and RCP, which reduces the recycling of α5β1 and EGFR (Muller et al., 2009), also blocked MenaINV-driven 3D haptotaxis and ECM reorganization. Whether or not Mena isoforms directly contribute to receptor cotrafficking is beyond the scope of the present study and will be the topic of future investigation.
Overall our study shed light on how cell migration behavior is coordinated by diverse and often overlapping signaling pathways that are activated during local tumor cell invasion, summarized in Figure 7. We focused on EGF and FN, both of which are present in the tumor microenvironment and can elicit directional tumor cell migration. However, given that there are many other cues present in the tumor microenvironment, it is likely that additional pathways participate in GF/ECM cross-talk downstream of a variety of cues and will contribute to metastasis.
FIGURE 7:
Model for MenaINV-driven cross-talk between EGFR and α5β1. MenaINV-driven chemotaxis requires the dysregulation of PTP1B, which leads to increased phosphorylation of both EGFR and paxillin located at FXs. MenaINV-driven haptotaxis requires EGFR-α5β1 cross-talk: in two dimensions, via dysregulation of PTP1B signaling at FXs, and in three dimensions, via receptor recycling. As a result, tumor cells expressing MenaINV show increased invasion in response to both EGF and FN relative to each cue alone.
MATERIALS AND METHODS
Antibody reagents, growth factors, and inhibitors
The growth factor EGF was from Life Technologies (Carlsbad, CA). Antibodies were as follows: EGFR (555996; BD Biosciences, San Jose, CA), paxillin (610052; BD Biosciences), pPaxillin Y118 (44-722G; Life Technologies), and pEGFR1173 (Abcam, Cambridge, MA). Drugs were erlotinib (LC Labs, Woburn, MA), gefitinib (Peprotech, Rocky Hill, NJ), Cilengitide (Selleck Chemicals, Houston, TX), P1D6 α5 blocking antibody (DSHB, Iowa City, IA), and PTP1Bi (Millipore, Burlington, MA).
Cell culture
MDA-MB-231 cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured in DMEM with 10% serum and penicillin-streptomycin glutamine. Retroviral packaging, infection, and fluorescence-activated cell sorting (FACS) were performed as previously described (Gupton et al., 2012). Cell lines engineered to stably express Mena isoforms expressed 10- to 15-fold more protein than wild-type cell line (Oudin et al., 2016b). Knockdown of p53, RCP, and EGFR was performed using previously published siRNA sequences (obtained from Dharmacon, GE Lifesciences, Lafayette, CO; Muller et al., 2009) and transient transfection with Dharmafect4.
Three-dimensional in vitro chemotaxis assay
We coated 96-well plates with 3% bovine serum albumin (BSA; Sigma, St. Louis, MO) at 37°C for 3 h. MDA-MB-231 cells expressing the different Mena isoforms were plated in 2 mg/ml collagen I (BD Biosciences) with 10× DMEM and 1 N NaOH and increasing concentrations of soluble bovine FN (Sigma) at 600,000 cells/ml on ice. Plates were then spun down at 1000 rpm for 8 min and placed in the incubator at 37°C for collagen to polymerize. After 1 h, cells were treated with various growth factors. At 24 h later, gels were fixed with 4% paraformaldehyde and stained with the nuclear dye Yopro (Life Technologies). Images of the individual gels were captured over a 300-μm distance with 5-μm steps with a Nikon (Melville, NY) spinning-disk confocal microscope using a 10×/0.75 numerical aperture (NA) Phase Plan-Apochromat objective, an Innova 70C Spectrum laser, and a Hamamatsu Orca ER camera (Hamamatsu, Bridgewater, NJ). A custom-made Matlab (Natick, MA) program was used to quantify the proportion of cells that migrated >20 μm (Miller et al., 2013, 2015). Data were collected from at least three different experiments with at least three replicate wells in each experiment. Synergy was calculated by two-way ANOVA.
Protrusion assay
Collagen-coated glass-bottomed dishes (MatTek, Ashland, MA) were coated with 0.2% Matrigel in serum-free medium for 30 min at 37°C. Cells were seeded sparsely overnight and then serum starved for 4 h in Leibowitz’s L-15 medium (Life Technologies) with 0.35% BSA. Differential interference contrast images were acquired every 20 s for 10 min in an environmentally controlled microscope (TE2000; Nikon) with a 20× objective and a Photometrics CoolSNAP HQ camera. Growth factor/inhibitor solutions were added after 80 s. Cell areas were traced immediately before and 9 min after stimulation using ImageJ (National Institutes of Health, Bethesda, MD). Data shown are from individual cells (at least 60 overall) pooled from at least three separate experiments.
Western blot
Standard procedures were used for protein electrophoresis, Western blotting, and immunoprecipitation. MDA-MB-231 cells expressing different Mena isoforms were lysed in 25 mM Tris, 150 mM NaCl, 10% glycerol, 1% NP-40, and 0.5 M EDTA with protease Mini-Complete protease inhibitors (Roche, Indianapolis, IN) and a phosphatase inhibitor cocktail (PhosSTOP; Roche) at 4°C. Protein lysates were separated by SDS–PAGE, transferred to a nitrocellulose membrane, blocked with Odyssey Blocking Buffer (Li-Cor, Lincoln, NE), and incubated in primary antibody overnight at 4°C. Proteins were detected using Li-Cor secondary antibodies. Protein level intensity was measured with ImageJ, and data were pooled from at least three different experiments.
FACS
MDA-MB-231 cells expressing the different isoforms were trypsinized, resuspended in medium, and then incubated with a primary antibody in phosphate-buffered saline (PBS) and 5% medium for 30 min on ice. Next the cells were incubated with a species-appropriate, Alexa 647–tagged secondary antibody and then resuspended in PBS with 10 μg/ml propidium iodide. Samples were then analyzed on a FACSCalibur machine (BD Biosciences). Data were pooled from at least three separate experiments, with 10,000 cells analyzed per experiment.
Tumor formation and in vivo invasion assay
MDA-MB-231 cells (2 million per mouse in PBS and 20% collagen I) expressing different Mena isoforms were injected into the fourth right mammary fat pad of 6-wk-old female NOD-SCID mice (Taconic, Hudson, NY). The in vivo invasion assay was performed in four mice per condition as previously described, once tumors had reached 1 cm in diameter, at ∼8 wk after injection (Wyckoff, 2004). Briefly, needles were held in place by a micromanipulator around a single mammary tumor of an anesthetized mouse. Needles contained a mixture of 0.5 mg/ml collagen I, EDTA with L-15 medium, and 5 nM EGF and/or 50 μg/ml FN. After 4 h of cell collection, the contents of the needles were extruded. Cells were then stained with 4′,6-diamidino-2-phenylindole and counted.
Haptotaxis assay
Microfluidic devices were prepared as previously described (Wu et al., 2012). For 2D haptotaxis, after bonding polydimethylsiloxane devices to MatTek dishes, the cell culture chamber was coated with 0.1 mg/ml collagen I for 1 h at 37°C, and then 250 μg/ml fluorescently labeled FN was flowed through the source channel for 1 h. Cells were then plated in the device in full serum medium and left to attach for 1 h before imaging. For 3D haptotaxis, cells were resuspended in 1 mg/ml collagen I (BD Biosciences) with 10× DMEM, 1 N NaOH, and 3 nM EGF, plated in the cell culture chamber, and left to settle for 8 h at 37°C. Fluorescently labeled FN (250 μg/ml unless otherwise specified) was then flowed through the source channel for 1 h before imaging.
Cells were imaged overnight in the haptotaxis device, with images acquired every 10 min for 16 h in an environmentally controlled microscope (TE2000, Nikon) with a 20× objective and a Photometrics CoolSNAP HQ camera. Individual cells were manually tracked using the ImageJ Manual Tracking plug-in. The tracks obtained were further analyzed using the chemotaxis tool developed by ibidi in ImageJ. This analysis tool was used to extract the FMI (Supplemental Figure S1A), which is defined as the displacement of the cell in the direction of the gradient divided by the total distance migrated; typical values for MDA-MB-231 haptotaxis on FN gradients were in the range 0.1–0.25. Note that the dimethyl sulfoxide (DMSO) control data for the MDA-MB-231 cells are the same for Figures 1C and 4G, given that both erlotinib and PTP1Bi are in DMSO and experiments were carried out at once.
Immunofluorescence
Cells were plated in a haptotaxis device on a 125 μg/ml 2D FN gradient for 3 h or on collagen-coated glass-bottomed dishes (MatTek) in serum-free medium for 30 min at 37°C. Cells were then fixed for 20 min in 4% paraformaldehyde in PHEM buffer (60 mM piperazine-N,N′-bis(ethanesulfonic acid), pH 7.0, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.0, 10 mM ethylene glycol tetraacetic acid, pH 8.0, 2 mM MgCl2, and 0.12 M sucrose) and then permeabilized with 0.2% Triton X-100, blocked with 10% BSA, and incubated with primary antibodies overnight at 37°C. Z-series of images were taken on an Applied Precision DeltaVision microscope using Softworx acquisition, an Olympus 40×/1.3 NA Plan Apo objective, and a Photometrics CoolSNAP HQ camera. Images were deconvolved using DeltaVision Softworx software and objective-specific point spread function. Images were analyzed with ImageJ. Images are pooled from at least three independent experiments, with at least 10 cells per experiment.
TCGA data analysis
Data retrieval from TCGA was explained in Oudin et al. (2016b).
Supplementary Material
Acknowledgments
We thank the Microscopy, Flow Cytometry, and Histology facilities in the KI Swanson Biotechnology Center for support. This work was supported by Department of Defense Breast Cancer Research Program W81XWH-12-1-0031 and a Ludwig Center for Molecular Oncology Postdoctoral Fellowship to M.J.O., funds from the Ludwig Center at the Massachusetts Institute of Technology to F.B.G., National Institutes of Health Grant U54-CA112967 to F.B.G. and D.A.L., the Koch Institute Frontier Research Program through the Kathy and Curt Marble Cancer Research Fund to F.B.G., and Koch Institute NCI Core Grant P30-CA14051.
Abbreviations used:
- ECM
extracellular matrix
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- FN
fibronectin
- GF
growth factor
- HGF
hepatocyte growth factor
- IGF
insulin growth factor
- PTP1Bi
inhibition of PTP1B
- RCP
Rab-coupling protein
- RTK
receptor tyrosine kinase
- TCGA
The Cancer Genome Atlas.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-04-0212) on August 24, 2016.
REFERENCES
- Astrof S, Crowley D, George EL, Fukuda T, Sekiguchi K, Hanahan D, Hynes RO. Direct test of potential roles of EIIIA and EIIIB alternatively spliced segments of fibronectin in physiological and tumor angiogenesis. Mol Cell Biol. 2004;24:8662–8670. doi: 10.1128/MCB.24.19.8662-8670.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanis N, Carlin CR. Mutual cross-talk between fibronectin integrins and the EGF receptor: molecular basis and biological significance. Cell Logist. 2012;2:46–51. doi: 10.4161/cl.20112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci. 2009;122:1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Haugh JM. Directed migration of mesenchymal cells: where signaling and the cytoskeleton meet. Curr Opin Cell Biol. 2014;30:74–82. doi: 10.1016/j.ceb.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdisso JE, González Á, Arregui CO. PTP1B promotes focal complex maturation, lamellar persistence and directional migration. J Cell Sci. 2013;126:1820–1831. doi: 10.1242/jcs.118828. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol. 2008;183:143–155. doi: 10.1083/jcb.200804140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KT, Asokan SB, King SJ, Bo T, Dubose ES, Liu W, Berginski ME, Simon JM, Davis IJ, Gomez SM, et al. LKB1 loss in melanoma disrupts directional migration toward extracellular matrix cues. J Cell Biol. 2014;207:299–315. doi: 10.1083/jcb.201404067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMilla PA, Stone JA, Quinn JA, Albelda SM, Lauffenburger DA. Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength. J Cell Biol. 1993;122:729–737. doi: 10.1083/jcb.122.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler F, Condeelis J. Metastasis: tumor cells becoming MENAcing. Trends Cell Biol. 2011;21:81–90. doi: 10.1016/j.tcb.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami S, Philippar U, Sun D, Patsialou A, Avraham J, Wang W, Di Modugno F, Nistico P, Gertler FB, Condeelis JS. Identification of invasion specific splice variants of the cytoskeletal protein Mena present in mammary tumor cells during invasion in vivo. Clin Exp Metastasis. 2009;26:153–159. doi: 10.1007/s10585-008-9225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Riquelme D, Hughes-Alford SK, Tadros J, Rudina SS, Hynes RO, Lauffenburger D, Gertler FB. Mena binds α5 integrin directly and modulates α5β1 function. J Cell Biol. 2012;198:657–676. doi: 10.1083/jcb.201202079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SK, Oudin MJ, Tadros J, Neil J, Del Rosario A, Joughin BA, Ritsma L, Wyckoff J, Vasile E, Eddy R, et al. PTP1B-dependent regulation of receptor tyrosine kinase signaling by the actin-binding protein Mena. Mol Biol Cell. 2015;26:3867–3878. doi: 10.1091/mbc.E15-06-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher A, Horwitz AR. Integrins in cell migration. Cold Spring Harb Perspect Biol. 2011;3:1–16. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HE, King SJ, Asokan SB, Rotty JD, Bear JE, Haugh JM. F-actin bundles direct the initiation and orientation of lamellipodia through adhesion-based signaling. J Cell Biol. 2015;208:443–455. doi: 10.1083/jcb.201406102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwada SK, Li X. Integrin alpha5/beta1 mediates fibronectin-dependent epithelial cell proliferation through epidermal growth factor receptor activation. Mol Biol Cell. 2000;11:2485–2496. doi: 10.1091/mbc.11.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AS, Miller Ma, Gertler FB, Lauffenburger Da. The receptor AXL diversifies EGFR signaling and limits the response to EGFR-targeted inhibitors in triple-negative breast cancer cells. Sci Signal. 2013;6:ra66. doi: 10.1126/scisignal.2004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Meyer AS, Beste MT, Lasisi Z, Reddy S, Jeng KW, Chen CH, Han J, Isaacson K, Griffith LG, Lauffenburger DA. ADAM-10 and -17 regulate endometriotic cell migration via concerted ligand and receptor shedding feedback on kinase signaling. Proc Natl Acad Sci USA. 2013;110:E2074–E2083. doi: 10.1073/pnas.1222387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Moss ML, Powell G, Petrovich R, Edwards L, Meyer AS, Griffith LG, Lauffenburger DA. Targeting autocrine HB-EGF signaling with specific ADAM12 inhibition using recombinant ADAM12 prodomain. Sci Rep. 2015;5:15150. doi: 10.1038/srep15150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra AK, Sawada K, Tiwari P, Mui K, Gwin K, Lengyel E. Ligand-independent activation of c-Met by fibronectin and α(5)β(1)-integrin regulates ovarian cancer invasion and metastasis. Oncogene. 2011;30:1566–1576. doi: 10.1038/onc.2010.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- Muller PA, Trinidad AG, Timpson P, Morton JP, Zanivan S, van den Berghe PV, Nixon C, Karim SA, Caswell PT, Noll JE, et al. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene. 2013;32:1252–1265. doi: 10.1038/onc.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudin MJ, Hughes SK, Rohani N, Moufarrej MN, Jones JG, Condeelis JS, Lauffenburger DA, Gertler FB. Characterization of the expression of the pro-metastatic Mena(INV) isoform during breast tumor progression. Clin Exp Metastasis. 2016a;33:249–261. doi: 10.1007/s10585-015-9775-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudin MJ, Jonas O, Kosciuk T, Broye LC, Guido BC, Wyckoff J, Riquelme D, Lamar JM, Asokan SB, Whittaker C, et al. Tumor cell-driven extracellular matrix remodeling enables haptotaxis during metastatic progression. Cancer Discov. 2016b;6:516–531. doi: 10.1158/2159-8290.CD-15-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul NR, Jacquemet G, Caswell PT. Endocytic trafficking of integrins in cell migration. Curr Biol. 2015;25:R1092–R1105. doi: 10.1016/j.cub.2015.09.049. [DOI] [PubMed] [Google Scholar]
- Roussos ET, Wang Y, Wyckoff JB, Sellers RS, Wang W, Li J, Pollard JW, Gertler FB, Condeelis JS. Mena deficiency delays tumor progression and decreases metastasis in polyoma middle-T transgenic mouse mammary tumors. Breast Cancer Res. 2010;12:R101. doi: 10.1186/bcr2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veevers-Lowe J, Ball SG, Shuttleworth A, Kielty CM. Mesenchymal stem cell migration is regulated by fibronectin through α5β1-integrin-mediated activation of PDGFR-β and potentiation of growth factor signals. J Cell Sci. 2011;124:1288–1300. doi: 10.1242/jcs.076935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wyckoff JB, Goswami S, Wang Y, Sidani M, Segall JE, Condeelis JS. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 2007;67:3505–3511. doi: 10.1158/0008-5472.CAN-06-3714. [DOI] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- Wyckoff JB, Segall JE, Condeelis JS. The collection of the motile population of cells from a living tumor. Cancer Res. 2000;60:5401–5404. [PubMed] [Google Scholar]
- Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- Zhou X, Rowe RG, Hiraoka N, George JP, Wirtz D, Mosher DF, Virtanen I, Chernousov MA, Weiss SJ. Fibronectin fibrillogenesis regulates three-dimensional neovessel formation. Genes Dev. 2008;22:1231–1243. doi: 10.1101/gad.1643308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Clark RAF. Fibronectin at select sites binds multiple growth factors and enhances their activity: expansion of the collaborative ECM-GF paradigm. J Invest Dermatol. 2014;134:895–901. doi: 10.1038/jid.2013.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.