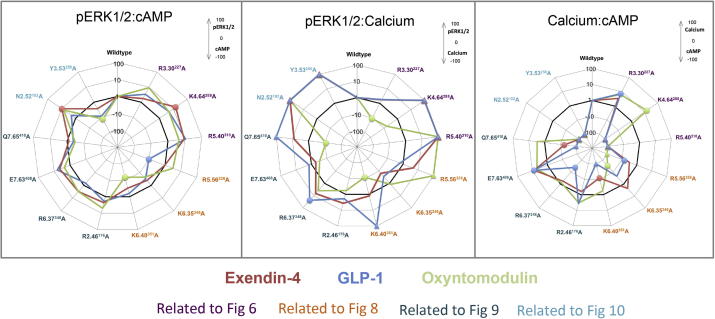
Fig. 5.
Effect of mutations on agonist bias of GLP-1R signalling pathways. Radial plots of agonist bias factors (ΔΔτ/KA, the ratio of the transduction coefficient for one pathway vs another, each normalised to the values determined for the wildtype receptor) derived from an operational model of agonism (see “Section 2”) plotted for each receptor variant. Values greater than 1 denote bias towards pathway 1, and values less than 1 denote bias towards pathway 2 relative to signalling at the wildtype receptor. Left, pERK1/2 (pathway 1) vs cAMP (pathway 2); middle, pERK1/2 (pathway 1) vs iCa2+ mobilisation (pathway 2); right, iCa2+ mobilisation (pathway 1) vs cAMP (pathway 2). All plots show the bias factors for the mutant receptors relative to the wildtype receptor for GLP-1 (blue), exendin-4 (salmon) and oxyntomodulin (green). Data points plotted as circles indicate statistically significant bias relative to the wildtype receptor (WT highlighted by the black reference line), whereas data plotted as triangles (at a value of −100 or 100) indicate that no significant signal could be detected for a particular pathway and therefor a bias factor could not be calculated. These values at −100 indicate no signalling in pathway 1 (therefore implied bias towards pathway 2), whereas +100 indicates no signalling in pathway 2 (therefore implied bias towards pathway 1). The residues are highlighted in the colour relevant to the clustering (and relevant figure) in which they are discussed in the results section.