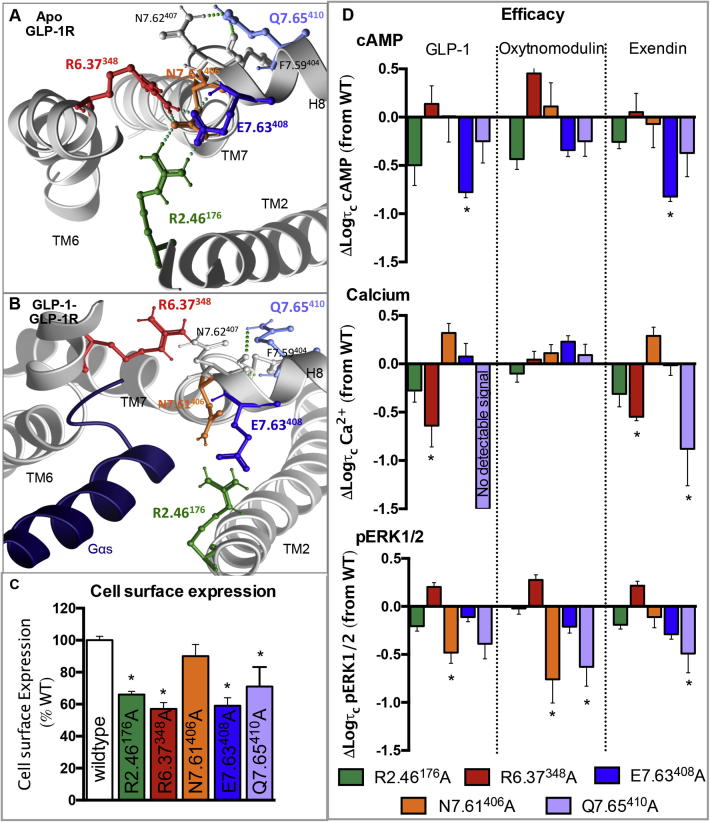
Fig. 9.
Effects on mutation of residues located in the hydrogen bonding network located between TMs 2, 6 and 7 at the cytoplasmic face. (A) TMs 2, 6, 7 and helix 8 (H8) of the apo GLP-1R TM bundle as viewed from the cytoplasmic face, highlighting an extensive hydrogen bond network between R2.46176, R6.37348, N7.61406 and E7.63408. Q7.65410 at the start of H8 is also shown where it forms hydrogen binds with the side chain of N7.61407 and the backbone of TM7 at F7.59404. (B) TMs 2, 6, 7 and H8 of the GLP-1 docked GLP-1R TM bundle as viewed from the cytoplasmic face with the Gαs peptide fragment indicating the extensive hydrogen bond network between R2.46176, R6.37348, N7.61406 and E7.63408 is broken in the activated receptor. Q7.65410 at the start of H8 is also shown where it still maintains a backbone interaction with F7.59404. (C) Cell surface expression of mutations R2.46176A, R6.37348A, N7.61406A, E7.63408A and Q7.65410A relative to the wildtype receptor (as assessed by antibody binding to the N-terminal c-myc epitope tag). (D) Differences in the coupling efficiency (log τc) of GLP-1, exendin-4 and oxyntomodulin to three signalling pathways (cAMP production (top), iCa2+ mobilisation (middle), and pERK1/2 (bottom)) for R2.46176A, R6.37348A, N7.61406A, E7.63408A and Q7.65410A compared to the wildtype receptor. These log τc were calculated from concentration–response curves presented in Fig. 2, Fig. 3, Fig. 4, and corrected for cell surface expression as measured by antibody labelling recorded in Table 1. Statistical significance of changes in cell surface expression or coupling efficacy in comparison with wildtype were determined by one-way analysis of variance and Dunnett’s post test, and values are indicated with an asterisk (∗, p < 0.05). All values are ±S.E.M of four to six independent experiments, conducted in duplicate.