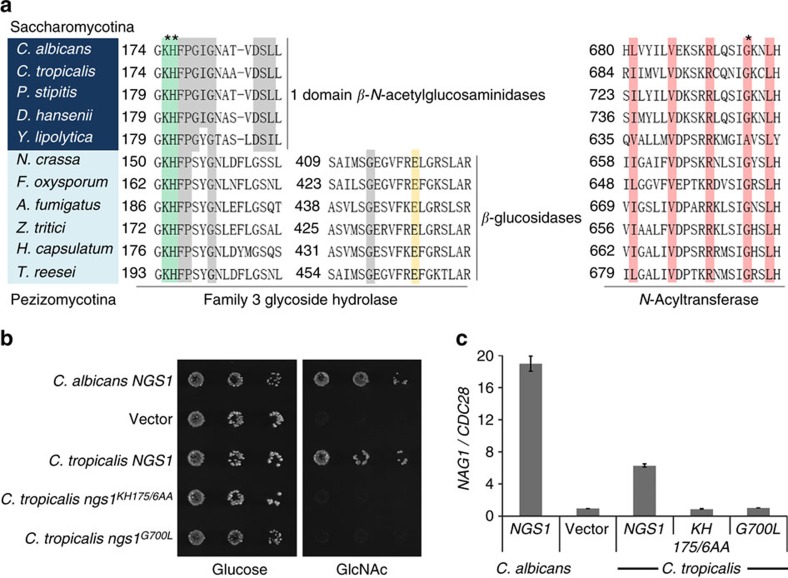
Figure 5. Evolutionary conservation of GlcNAc signalling in Ascomycota.
(a) Partial multiple sequence alignment of Ngs1 orthologs in the GH3 glycosidase domain and the N-Acyltransferase domain from Saccharomycotina (shaded in dark blue) and Pezizomycotina (shaded in light blue). The conserved residues in one domain β-N-acetylglucosaminidases and two domain β-glucosidases, as determined by Litzinger and Fischer29, are shaded in grey. The residues known to interact with GlcNAc (shaded in green) and the glutamate residue known to act as the acid/base catalyst in β–glucosidases subfamily (shaded in yellow) are indicated. Residues shaded in pink in the N-Acyltransferase domain are conserved in motif A, as shown in Fig. 4a. The asterisks indicate the positions used in the mutational analysis in C. tropicalis. (b) Ectopically expressed CtNGS1 suppresses the growth defect of Cangs1 mutant on GlcNAc. Dilutions of C. albicans ngs1 mutant cells carrying CaNGS1 (HLY4395), CtNGS1 (HLY4456), Ctngs1KH175/6AA (HLY4457), Ctngs1G700L (HLY4458) or vector alone were grown on YNB plates containing 2.5 mM GlcNAc or glucose at 30 °C for 2 days. (c) The conserved key GlcNAc binding sites K175H176 and acetyl-CoA interacting residue G700 in CtNgs1 are required for the induction of NAG1 in C. albicans. NAG1 mRNA levels of the strains from (b) were determined. Cells were grown in liquid SC medium supplemented with 2.5 mM GlcNAc at 30 °C and collected at 15 min for qRT-PCR analysis. Mean data±s.d. from three independent qRT-PCR experiments was plotted.