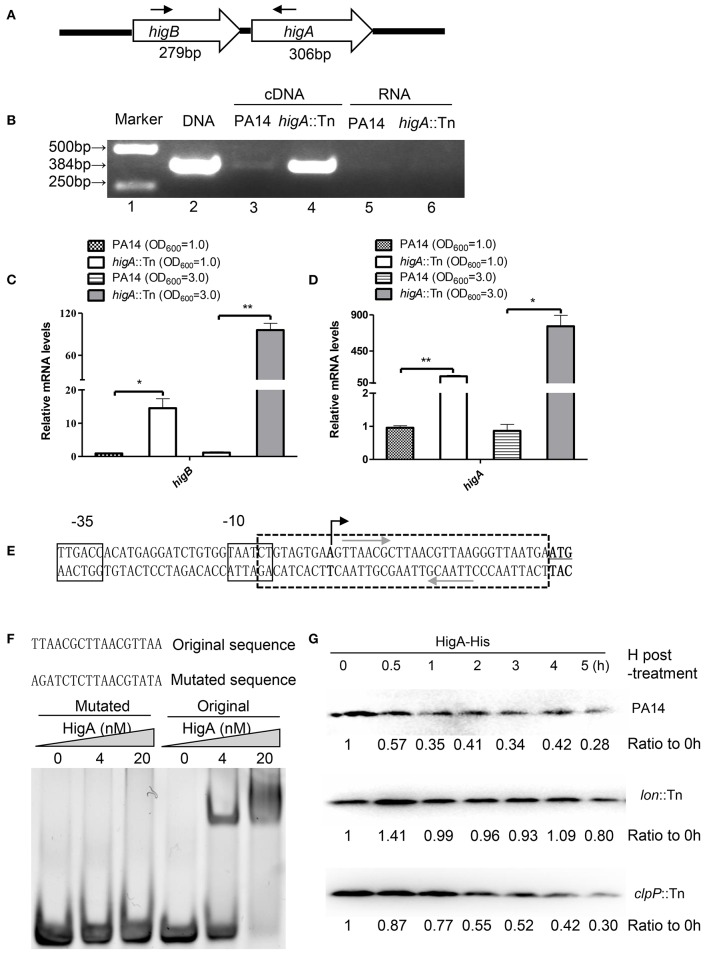
Figure 1.
HigA negatively regulates the higB-higA operon. (A) Sketch map of the higB-higA operon. Arrows indicate the directions and locations of the primers for RT-PCR. (B) Total RNA was isolated from PA14 and the higA::Tn mutant. cDNA was synthesized and used as templates in PCR. RNAs were used in RT-PCR as negative controls. (C,D) The relative mRNA levels of higB and higA genes in PA14 and the higA::Tn mutant. Total RNA was isolated and the mRNA levels were determined by quantitative RT-PCR with the 16S rRNA as the internal control. Data represents the mean ± standard deviation from three independent experiments performed in triplicate. *p < 0.05; **p < 0.01 compared to wild type PA14 by Student's t-test. (E) Promoter region of the higB-higA operon. The predicted −10 and −35 elements of the promoter are boxed. The transcriptional start site is indicated by a black arrow, and the start codon of higB is underlined. The palindromic sequences of hypothetical HigA binding sites are indicated by gray arrows. (F) EMSA displaying binding of HigA to the higB-higA promoter. Purified HigA-His protein was incubated with the 38-bp DNA fragment indicated by the box with dashed lines in (E) or altered sequence. The mixtures were electrophoresed and observed by ethidium bromide staining. (G) Cleavage of HigA by the Lon protease. Wild type PA14, the clpP::Tn and lon::Tn mutants carrying pMMB67EH-higA-His were cultured in the presence of 1 mM IPTG for 1 h. Then 50 μg/ml spectinomycin was added to the medium. At indicated time points, the HigA-His levels were determined by Western blot analysis with an anti-His antibody. The relative density of each band was determined with Image J.