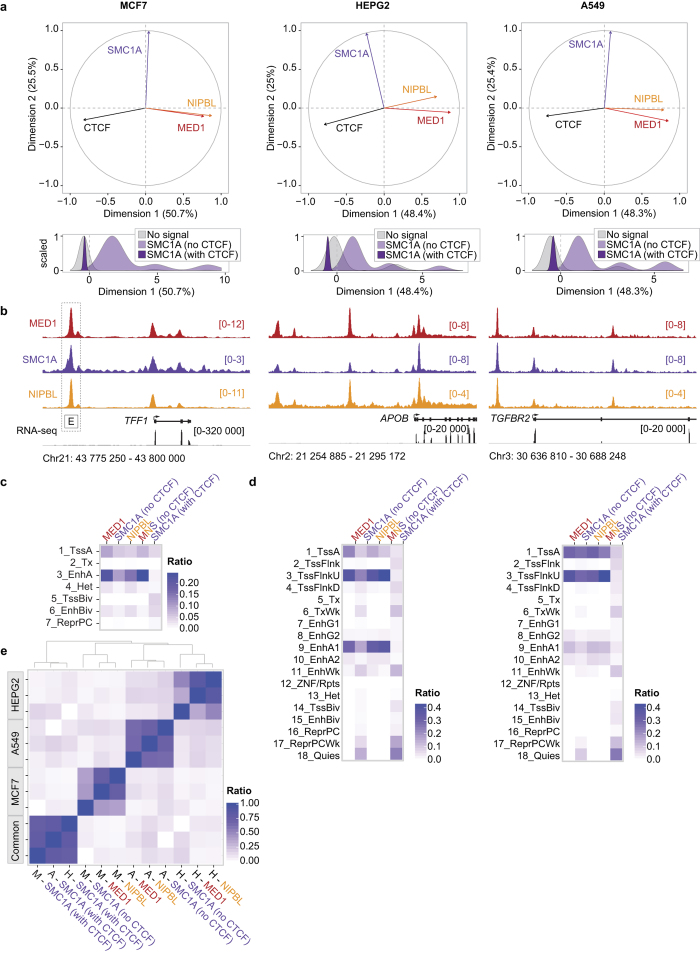
Figure 1. Mediator, Cohesin and NIPBL co-localize at regulatory regions of actively transcribed genes in cancer cells.
(a) Principal component analysis (PCA) of the distribution of MED1-, SMC1A-, NIPBL- and CTCF-occupied regions. Top – PCA variable factor map of the MED1, SMC1A, NIPBL and CTCF variables on the first two components showing a positive correlation between NIPBL and MED1. In addition, dimension 1 captures the negative correlation between the transcription factor CTCF and MED1/NIPBL-occupied regions. Bottom – Density plot of the genomic region coordinates on the dimension 1 showing a split distribution of SMC1A. SMC1A is associated with CTCF (negative values) or MED1 and NIPBL (positive values). The y axes are normalized to 1 while the x axes represent arbitrary units. (b) Occupancy profiles of Mediator (MED1), Cohesin (SMC1A) and NIPBL showing colocalization at regulatory regions. The TFF1, APOP and TGFBR2 loci are displayed for MCF7, HEPG2 and A549 respectively. The characterized enhancer region of TFF1 (box labeled E) is indicated60. The scales of ChIP-Seq and RNA-Seq profiles are displayed in reads per million. (c) Overlaps between MED1, SMC1A (no CTCF), SMC1A (with CTCF), the regions co-occupied by MED1-SMC1A-NIPBL and the functional annotation of the MCF7 genome (see Supplementary Information). MED1, SMC1A (no CTCF) and NIPBL occupy active TSS (1_TssA) and active enhancers (3_EnhA), but not inactive regions. The color scale indicates the ratio of overlap. (d) Overlaps between MED1, SMC1A (no CTCF), SMC1A (with CTCF), the regions co-occupied by MED1-SMC1A-NIPBL and the functional annotation of the HEPG2 (left) and A549 (right) genomes using the ChromHMM 18-state model32,33. MED1, SMC1A (no CTCF) and NIPBL occupy active promoters (1_TssA and 3_TssFlnkU) and active enhancer regions (8_EnhG2, 9_EnhA1 and 10_Enh_A2), but not inactive regions in both HEPG2 and A549 cells (see Supplementary Information for abbreviations). The color scales indicate the ratio of overlap. (e) Heat map showing the colocalization frequencies of MED1, SMC1A and NIPBL enriched regions in MCF7, HEPG2 and A549 cells. The color scale reflects the colocalization of each pair of regulators. Regulators were clustered along both axes based on the similarity in their correlation with each other.