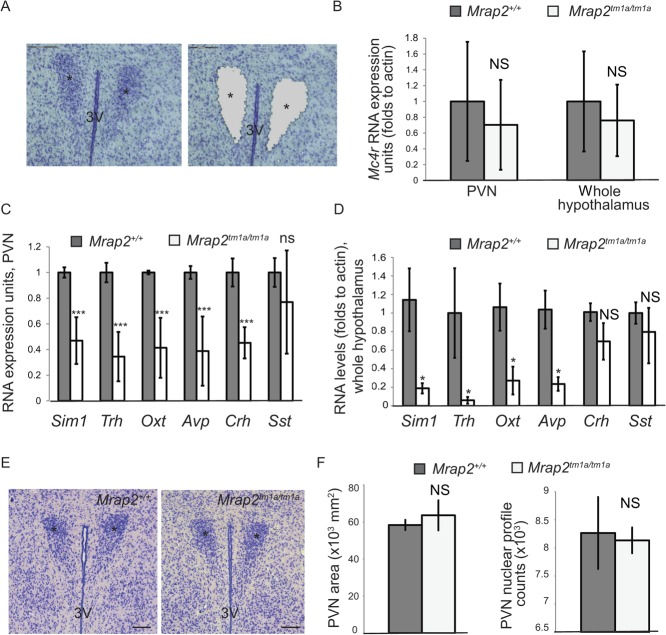
Figure 5.
Mrap2 is involved in Mc4r regulation in the hypothalamus. (A) An example of the hypothalamic section stained with cresyl violet before the microdissection (left panel) and after (right panel). Asterisks show the position of the PVN, 3 V-third ventricle, the scale bars are 150 μm. (B) Mc4r expression level in the PVN (Mrap2+/+ n=3, Mrap2tm1a/tm1a n=3) and in the whole hypothalamus (Mrap2+/+ n=4, Mrap2tm1a/tm1a n=4) as determined by the qPCR. (C) Expression of Sim1, Trh, Oxt, Avp, Crh and Sst in the PVN of 129/Sv wild type (n=4) and Mrap2tm1a/tm1a (n=3) mice. The data are represented as the mean of the microarray fluorescence values (±s.e.m.), normalized to the wild type for each gene. *P<0.05; **P<0.005; ***P<0.0005. (D) Expression of Sim1, Trh, Oxt, Avp, Crh and Sst in the whole hypothalamus of the wild-type and Mrap2tm1a/tm1a mice as determined by the qPCR. Data from male mice n=4 per genotype is shown. (E) Morphology of the PVN of 129/Sv Mrap2tm1a/tm1a mice (right panel) compared with the wild type as shown by representative images of coronal brain sections (approx. bregma –0.8 mm) stained by Nissl. (F) Average PVN area size (left graph) and stereotaxic counts of Nissl positive cells (right graph) in the PVN of the mutant 129/Sv mice (n=3) and their wild-type littermates (n=3).

 This work is licensed under a
This work is licensed under a