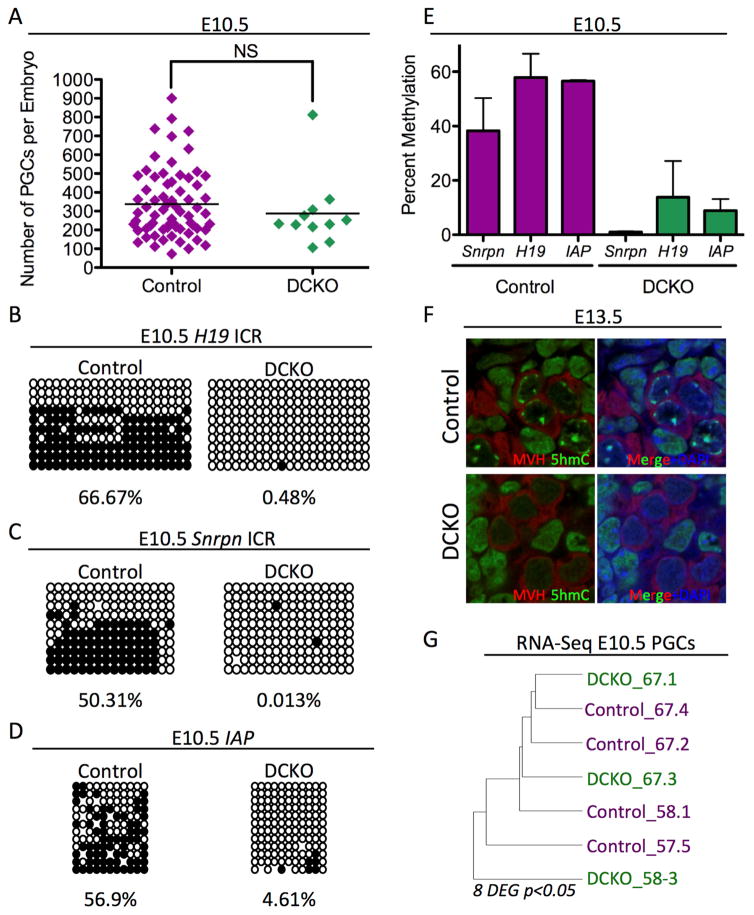
Figure 1. Dnmt1 Conditional knockout PGCs are hypomethylated.
(A) Total umber of PGCs sorted at E10.5. Error bars, ± SEM; NS: p=0.3986, two-tailed unpaired t test; n =78 biological replicates. (B–D) Bisulfite PCR of sorted PGCs from Control and DCKO embryos at E10.5 evaluating the H19 ICR (B), Snrpn ICR (C) and IAP EZ (D). For each locus at least 20 clones were sequenced. (E) Graphical representation of Bisulfite PCR methylation levels found in respective loci from cells at E10.5 n=2 biological replicates. (F) Immunofluorescence of E13.5 DCKO and Control male gonads evaluating MVH positive PGCs (red), 5hmC (green), and DAPI (blue) (n=3). (G) Unsupervised hierarchical clustering RNA-Seq analysis of PGCs at E10.5. p<0.05 and FDR of 10%. DEG = Differentially expressed genes. See also Figure S1