Figure 1.
Structure- and Design-Based Approaches for Identifying RNA-Binding Molecules
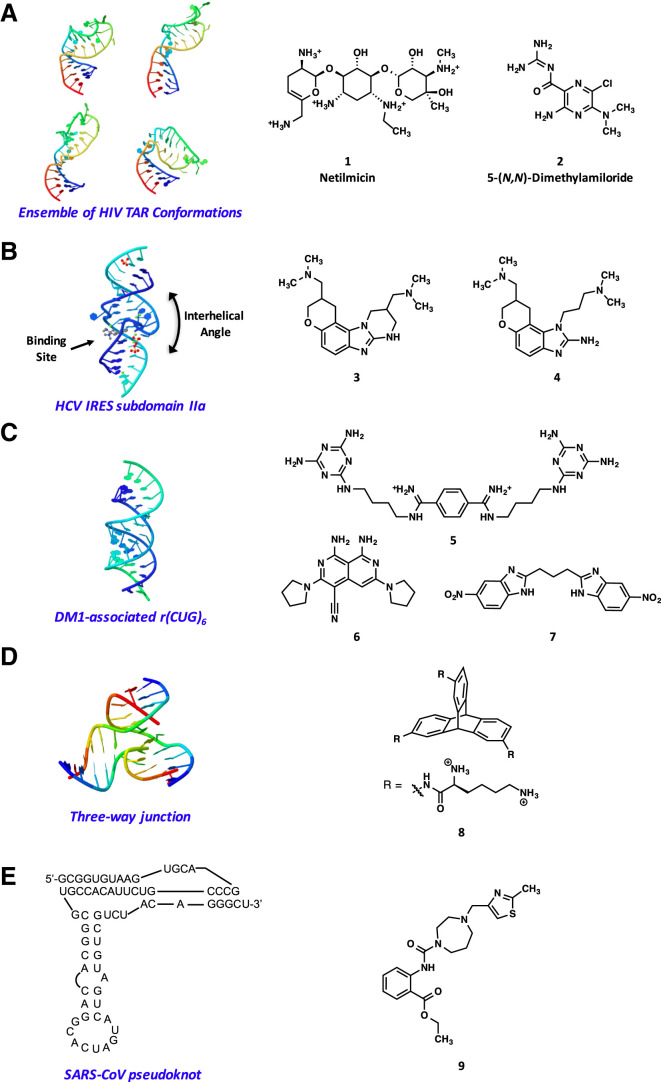
(A) Representative conformers of wild-type HIV-1 TAR RNA (left) used to identify TAR-binding small molecules (right) by virtual screening (Stelzer et al., 2011).
(B) Crystal structure of the HCV IRES subdomain IIa RNA in complex with the benzimidazole inhibitor 4 showing the small-molecule-induced change in the interhelical angle of the RNA (left), and structures of the identified HCV translation inhibitors that bind to subdomain IIa (right) (Parsons et al., 2009).
(C) Structure of the r(CUG)6 duplex (left) used to rationally design a highly selective CUG binding ligand 5 based on the triaminotriazine scaffold (right) (Nguyen et al., 2015, Wong et al., 2014). Structures of r(CUG)exp binding compounds 6 and 7 identified by high-throughput screening efforts are also shown (right) (Childs-Disney et al., 2013, Rzuczek et al., 2015).
(D) Representative structure of a three-way junction (left) for which RNA-binding triptycene-based scaffolds such as compound 8 (right) were developed (Barros et al., 2016a).
(E) Structure of the SARS-CoV RNA pseudoknot involved in an essential −1 ribosomal frameshift (left) used in a virtual screen to identify a SARS-pseudoknot-binding ligand (right) (Park et al., 2011).