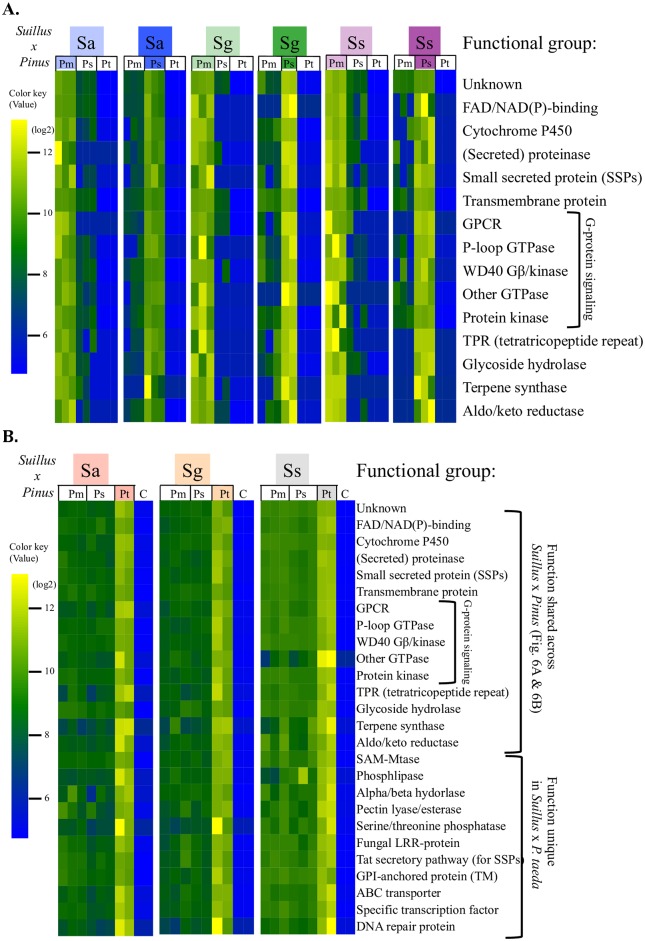
Fig 5. Comparative gene expression of Suillus genes expressed with different Pinus hosts during compatible (Fig 6A) and incompatible (Fig 6B) mycorrhizal interactions.
Suillus genes identified from Fig 4 were grouped according to function and relative expression rate (SI text A4), and plotted as a heatmap (color coding same as in Fig 4). Suillus/Pinus species pairs with over 500 unique genes are shown in Fig 4A. Only 10 unique genes were identified from S. decipiens/Pinus pairs. These include 2 SSPs, 1 P-loop, 1 WD40 and 6 unknown; their annotations are provided (with other 3 Suillus spp. in S1 Dataset. Significance was determined by normalization of reads across pairings (using DESeq) with false discovery rate (FDR) of 5% using Benjamini-Hochberg test to identify highly expressed transcripts with at least 2-fold change. The color key shows log2 fold changes of the normalized read number. Gene expression in uninoculated P. taeda roots (“C”) is also shown. Complete read count data for all genes and treatments are shown in S1 Dataset.