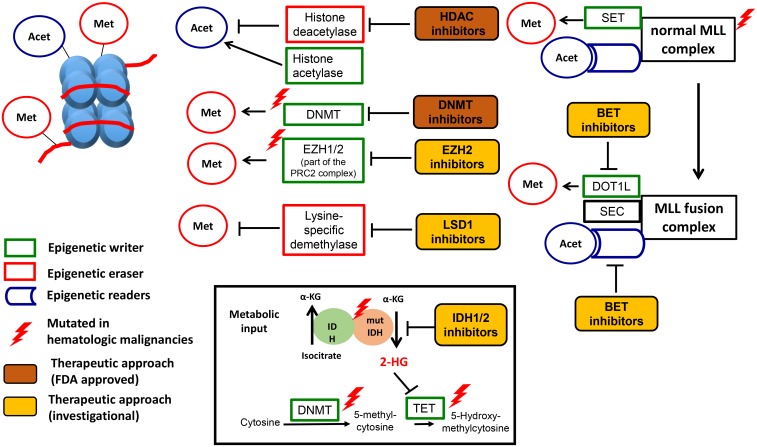
Fig 1. Epigenetics in hematological malignancies.
Epigenetic regulation, dysregulation and therapeutic targets. DNA (red) forms a complex with histone proteins (light blue) to form nucleosomes. Each nucleosome consists of DNA wrapped around a unit of eight histone proteins. Epigenetic regulation: Epigenetic marks are placed both on the DNA and histones by epigenetic writers (in green). DNMT3A, TET2, EZH1/2, and the histone acetylase are examples of epigenetic writers. EZH2 is part of the Polycomb Repressive Complex 2 (PRC2), which also contains ASXL1, EED, SUZ12, and RBAP48. Epigenetic marks are removed by epigenetic erasers (in red), e.g., Lysine specific demythylases (LSD) and histone deacetylases (HDAC). IDH1/2 provide metabolic input by providing a-KG, which is an important substrate for the catalytic domain of other epigenetic regulators like TET2. Finally, epigenetic marks are recognized by epigenetic regulators though special reader domains (in blue), which lead to the recruitment of epigenetic regulators to DNA and histones. Examples of reader domains are the plant homeodomain (PHD) finger proteins and the bromodomain and extraterminal (BET) family of proteins. The BET family has four members, including bromodomain-containing proteins 2, 3, and 4 (BRD2, BRD3, and BRD4) and BRDT. The wild-type mixed-lineage leukemia (MLL) gene is post-translationally cleaved into N-terminal and C-terminal fragments that re-associate to form the MLL complex. The C-terminal fragment contains a SET domain, which methylates H3K4, and the N-terminal fragment contains PHD fingers and a bromodomain, which serve as reader domains. Epigenetic dysregulation: The complex epigenetic regulatory program is disturbed in hematologic malignancies by mutations in epigenetic regulators (indicated by red thunderbolt) or by the recruitment of large multi-protein complexes like the MLL fusion complex (purple circle). Translocations involving the MLL gene account for the vast majority of infantile and approximately 10% of adult leukemias. Following translocation, a fragment of the N-terminal portion of MLL is fused in frame with a translocation partner, leading to the formation of novel MLL-fusion protein complexes, including the super elongation complex (SEC) and the DOT1-Like Histone H3K79 Methyltransferase (DOT1L) complex. The DOT1L complex leads to misdirected H3K79 methylation, which has been shown to sustain the expression of key pro-leukemic genes such as the HOXA genes and MEIS1. The SEC complex phosphorylates RNA polymerase II (POL II) facilitating its recruitment to the promoters of crucial oncogenes such as MYC, BCL2, and CDK6. Metabolic dysregulation is caused by IDH1/2 mutations, which leads to the production of an abnormal metabolite in the cell, 2-hydroxyglutarate (2HG), which can inhibit the hydroxylation of 5-mC by TET2. Therapeutic targeting: Food and Drug Administration (FDA)-approved are DNMT3A inhibitors for AML and MDS and HDAC inhibitors for T cell lymphoma and multiple myeloma, respectively (in orange). Several investigational drugs (in yellow) are in different stages of preclinical and clinical development. Adopted from Semin Hematol. 2015 Jul;52(3):172–83 [8]