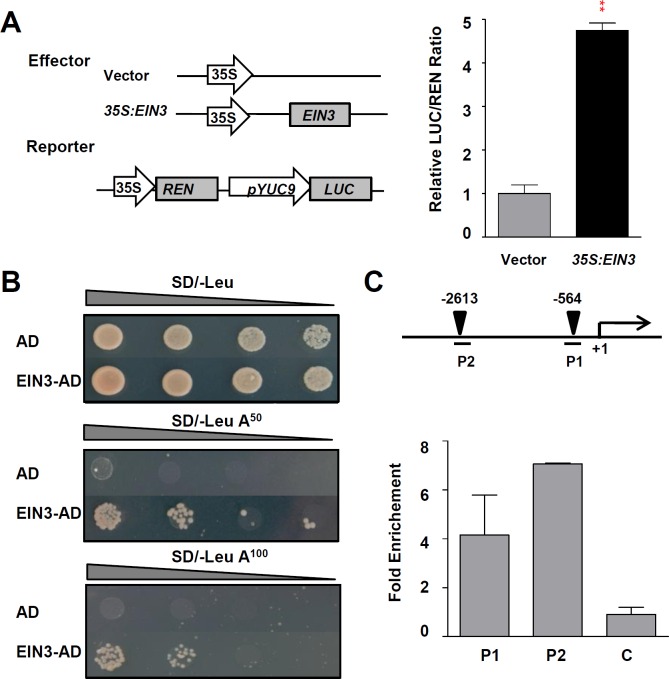
Fig 5. EIN3 is a transcriptional activator of YUC9.
(A) Left panel: Schematic diagrams of effector and reporter constructs used in the transient dual-luciferase assays. CaMV 35S promoter driving EIN3 (35S:EIN3) was used as effector, and empty vector as a negative control. Promoter fragments of YUC9 were used to make the YUC9p-LUC reporter. Right panel: Transient dual-luciferase assay shows that EIN3 transactivates the promoter of YUC9 in Arabidopsis protoplasts. Data represent the means of three biological replicates. Error bars represent Student’s t test confidence intervals (n ≥ 9). Statistical significant difference indicated by asterisks (Fisher’s exact test, **P < 0.01). (B) Physical interactions of EIN3 with YUC9 promoter in Y1H assays. Yeast expression plasmids pGADT7-EIN3 were reintroduced into yeast strain Y1H Gold carrying the reporter gene AbAr under the control of the YUC9 promoter. The transformants were screened for their growth on the yeast synthetic defined media (SD/-Leu) in the presence of 50 or 100 ng ml-1 Aureobasidin A (AbA) for stringent selection. The empty vector pGADT7 was included as a negative control. Yeast cultures were diluted (1:10 successive dilution series) spotted onto plates. (C) EIN3 associated with the promoter of YUC9 in ChIP-qPCR assay. Up panel: Schematic diagrams of YUC9 promoter of showing the potential EIN3 binding site (black triangles). The translational start sites (ATG) are shown as +1. Numbers above the diagram indicated the distance away from ATG. DNA fragments (P1 and P2) were used for ChIP. Chromatins isolated from 35S:EIN3-GFP transgenic line and 35S:GFP control were immunoprecipitated with anti-GFP antibody followed by qPCR to amplify P1 and P2 regions. Segment C located in the coding region was used as negative control. Input sample was used to normalize the qPCR results in each ChIP. Fold enrichment was presented as a ratio of normalized results from 35S:EIN3-GFP plants and 35S:GFP. Data are mean ± SD.