Fig. 3. Interprotein structure factor and its correction to obtain single-protein dynamics.
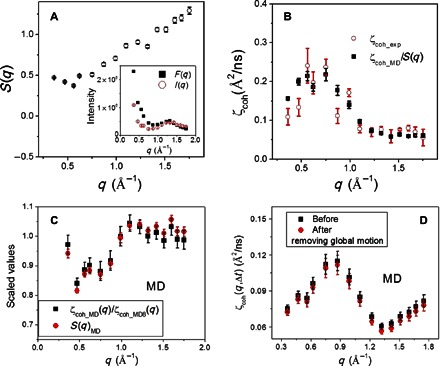
(A) Experimental interprotein structure factor, S(q) = I(q)/F(q). To obtain S(q) on an absolute scale, I(q) was multiplied by a constant to match the values of F(q) at q > 1 Å−1 (inset), because the interprotein scattering at high q is much smaller than at low q (32). (B) Comparison of ζcoh_exp(q,Δt) with ζcoh_MD(q,Δt) corrected by experimental S(q). (C) Comparison of MD-derived S(q)MD with the ratio between the single-protein ζcoh_MD(q) and the overall value, ζcoh_MD8(q). ζcoh_MD(q) is the average over the values calculated from each of the eight protein molecules in the simulation box, whereas ζcoh_MD8(q) is estimated by taking the eight protein molecules as a whole solution. S(q)MD arises from the interprotein packing in the MD box. (D) Comparison of ζcoh_MD(q,Δt) before and after removing the global motion of the protein molecule. Removal of the global motion was performed by fitting the MD trajectory of each protein molecule to its structure in the first frame.
