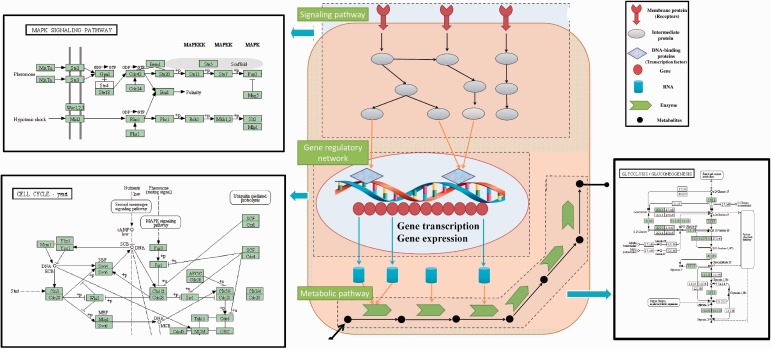
Figure 1.
Interrelationship among signaling pathway, gene regulatory network and metabolic pathway in a cell. Cell starts to recognize and receive the signals by activating the membrane receptors, responding to the stimulus of the changes in the outside environment. The receptors can help transmit the signal outside the cell membrane into the signaling pathway inside the cell, activating a series of biochemical reactions. Take the MAPK signaling pathway in yeast (Top-right) from KEGG [2] as example, the signaling pathway is initialized by the stimuli of peptide mating pheromones, which are Mat-alpha and MatA, then the receptor Ste2 or Ste3 connects the peptide mating pheromone and activates the MAPK signal transduction cascades. The signaling cascade will finally produce the protein kinases that can enter the nucleus and activate the gene transcription by binding the transcription factors onto promoters of DNA. In the example of cell cycle yeast pathway (Bottom-left), the Cln3-Cdc28 protein kinases activate the transcription factors SBF and MNF, which regulate the Cln1/2 gene expression. The gene regulatory network can control the gene expression levels of mRNA by activating the transcription factors, and further translate the mRNA into proteins. Specific proteins, called enzymes, will participate in the metabolic pathway and catalyze a series of biochemical reactions by converting substrates into products, in which the product of one reaction becomes the substrate of next reactions. In the Glycolysis/Gluconeogenesis pathway (Bottom-right), experiments [3] showed the regulators identified in cell cycle also regulated the metabolic enzymes to catalyze the cellular metabolism. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)