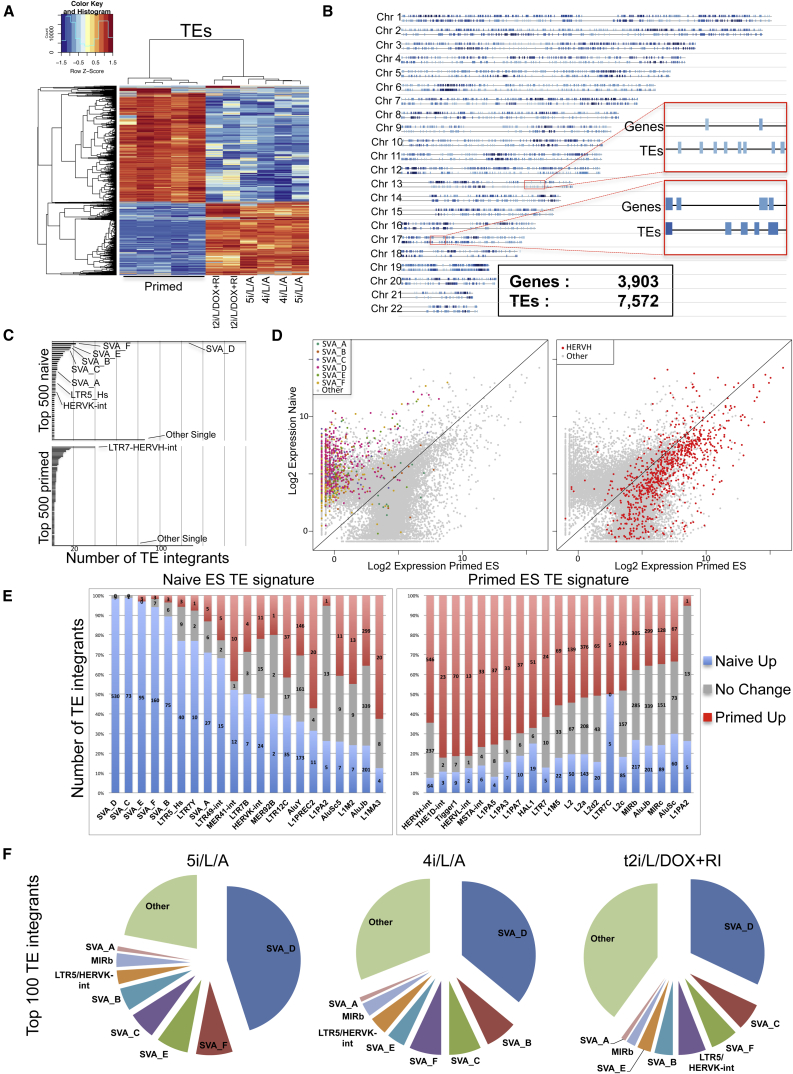
Figure 1.
TEs Accurately Discriminate Naive and Primed Human ESCs
(A) Heatmap of RNA-seq expression data from primed human ESCs and naive human ESCs derived in 5i/L/A-, 4i/L/A-, and transgene (KLF2/NANOG)-dependent naive cells maintained in t2i/L/DOX+RI. Data shown include 10,000 TEs with the highest SD between the samples.
(B) Naive/primed differentially expressed TEs (bottom) or genes (top) are depicted along human chromosomes. A cutoff fold change of 10× (p < 0.05) between naive and primed states was used to visualize the distribution of the most significantly differentially expressed TEs and genes. Note that TEs provide a much denser coverage of the genome.
(C) The top 500 TE integrants with the highest fold change between primed and naive ESCs are heavily enriched for HERVH-int-LTR7 (primed) or SVA and LTR5_Hs integrants (naive).
(D) Dot plot of all expressed TE integrants in naive versus primed ESCs. SVA-D, SVA-F, SVA-E, SVA-C, and SVA-B integrants are broadly expressed in naive ESCs (left), while SVA-A integrants are more evenly distributed. Similarly, HERVH-ints were by and large overexpressed in primed ESCs (right)
(E) TE signatures of naive or primed ESCs. The most heavily primed or naive-biased TE families are represented as columns split into three segments: naive-overexpressed integrants (2-fold cutoff, p adj. < 0.05), primed-overexpressed integrants, and no change (integrants with expression ratio not contained in the above cutoff).
(F) Pie charts comparing the top 100 TEs with highest fold change for different naive media (5i/L/A, 4i/L/A, and t2i/L/DOX+RI) compared to primed ESCs. The overall distribution is similar, with SVA-D representing about one-third of the top 100 TEs.
See also Figures S1 and S2.