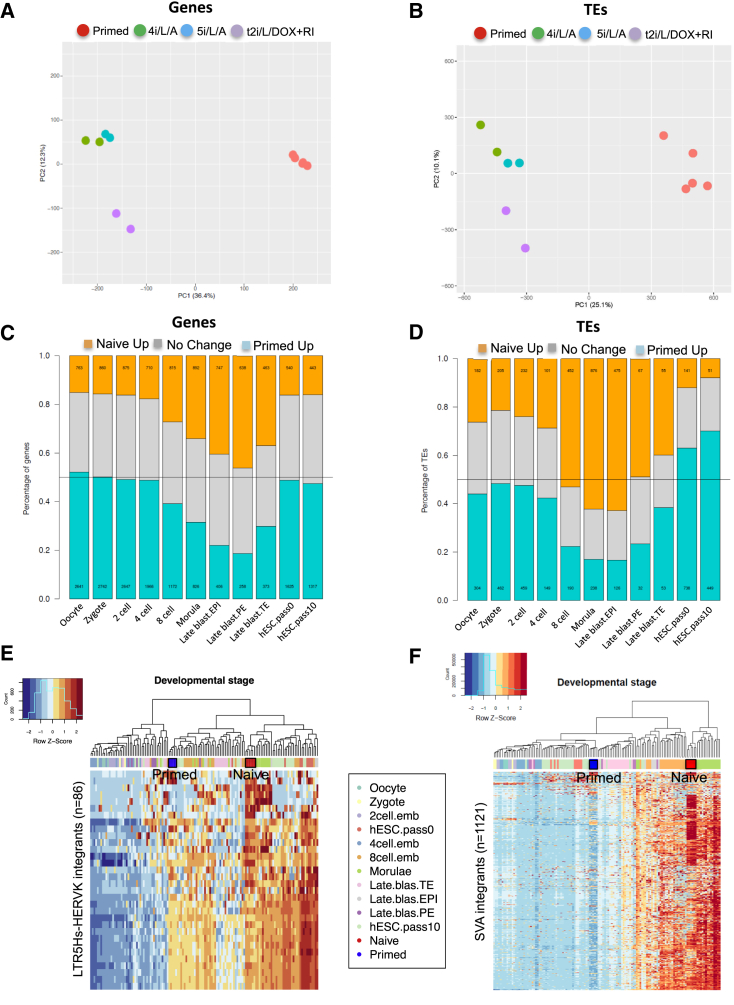
Figure 2.
Naive Human ESCs Have a Transposon Transcription Signature of the Human Preimplantation Embryo
(A and B) Principal component analysis (PCA) of primed or naive human ESCs derived in 5i/L/A-, 4i/L/A-, and transgene (KLF2/NANOG)-dependent naive cells maintained in t2i/L/DOX+RI based on the differential expression of genes (A) or transposable elements (B).
(C and D) Correspondence between gene expression (C) or TE expression (D) in naive/primed ESCs and single-cell human embryonic stages (Yan et al., 2013). For every stage of human embryonic development, a statistical test was performed to find the genes (or TEs) that have a different expression level compared to the other stages. The proportions of developmental stage-specific genes (or TEs) that are upregulated (p < 0.05, 2-fold change) in naive or primed cells are indicated in orange and blue, respectively, while genes (or TEs) that did not change expression are indicated in gray. The naive samples include all three conditions of naive cells that we examined in the PCA analyses (i.e., 5i/L/A, 4i/L/A, and t2i/L/DOX+RI). See Supplemental Experimental Procedures for details of the analysis.
(E and F) Heatmaps of RNA-seq data from naive/primed ESCs and from single-cell human embryonic stages (Yan et al., 2013). Clustering is performed with all LTR5_Hs/HERVK elements with expression data (n = 86) (E) or all SVA elements with expression data (n = 1121) (F). The naive samples shown in red include all three conditions of naive cells that we examined in the PCA analyses (i.e., 5i/L/A, 4i/L/A, and t2i/L/DOX+RI).
See also Figures S1–S3.