Figure 3.
Chromatin Expansion and Histone Mobilization on Chromatin Contribute to Parental H3.3 Redistribution after UVC Damage
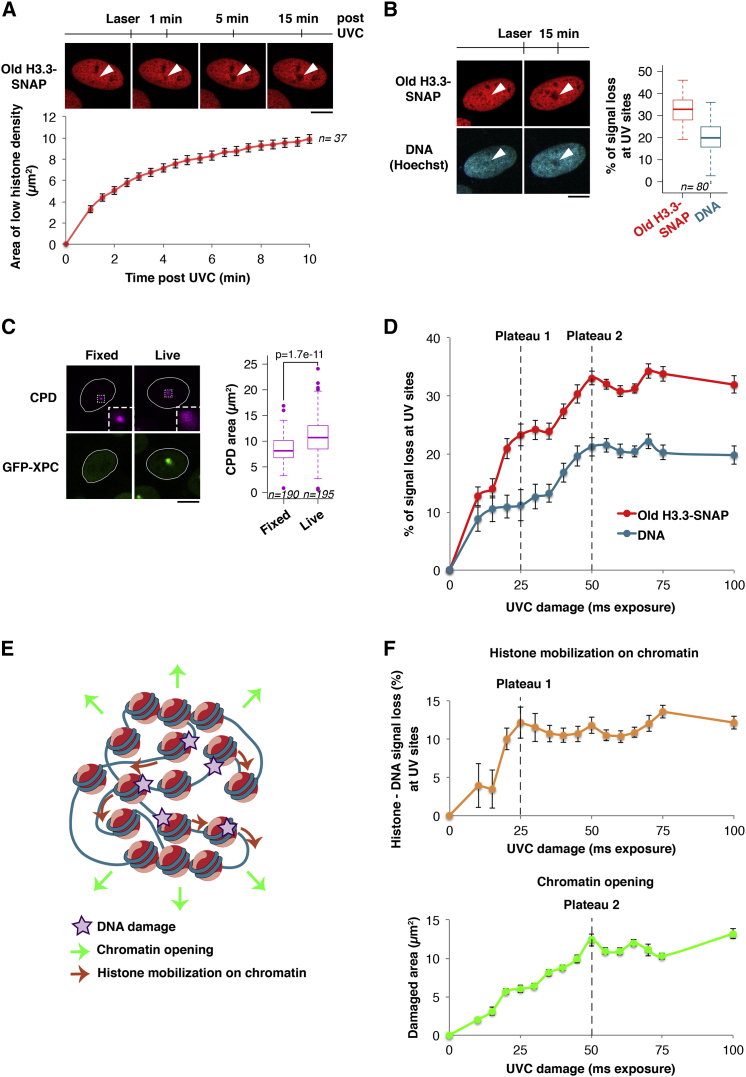
(A) Distribution of parental H3.3 (red) at the indicated time points after UVC laser damage in U2OS H3.3-SNAP cells. The area of parental histone loss is measured as a function of time post-UVC. Error bars, SEM from n cells scored in two independent experiments.
(B) Distribution of parental H3.3 (red) and DNA (blue, stained with Hoechst) before and 15 min after local UVC damage in U2OS H3.3-SNAP cells. The reduced Hoechst staining observed at damage sites is not due to photo-bleaching by the UVC laser (data not shown) or to DNA denaturation during UVC damage repair (Figure S3F). White arrowheads, irradiated areas. The boxplot shows quantifications of histone (red) and DNA (blue) fluorescence loss in irradiated areas (n cells scored in two independent experiments). Staining parental H3.3 in green and DNA in red with NUCLEAR-ID Red DNA stain gave similar results (data not shown).
(C) UVC-damaged chromatin areas visualized by staining for cyclobutane pyrimidine dimers (CPD, purple) after UVC laser damage in U2OS cells stably expressing H3.3-SNAP and GFP-XPC. The initial damaged chromatin area was measured in paraformaldehyde-fixed cells (no expansion, no GFP-XPC recruitment) and compared to 15 min after irradiation in live cells (n cells scored in two independent experiments). Insets: zoomed-in views (×3) of dashed line boxes. Scale bars, 10 μm.
(D) Quantification of parental H3.3 (red) and DNA (blue) fluorescence loss in irradiated areas as a function of UVC damage. The saturations of the two mechanisms contributing to parental histone loss (dotted lines) are obtained from the graphs in (F).
(E) Working model for parental H3.3 redistribution around UVC damage sites.
(F) Top: difference between parental H3.3 and DNA signal loss in irradiated areas as a function of UVC damage, which reflects histone mobilization on chromatin. Bottom: expansion of the damaged area marked by GFP-XPC as a function of UVC damage, indicative of chromatin opening. Dotted lines indicate the saturation of each mechanism. Error bars, SEM from at least 35 cells scored in three independent experiments.
See also Figures S3 and S5.