Figure 5.
Reversing Condensin-Dependent Overwinding Allows Top2-Dependent SCI Formation in Metaphase Cells
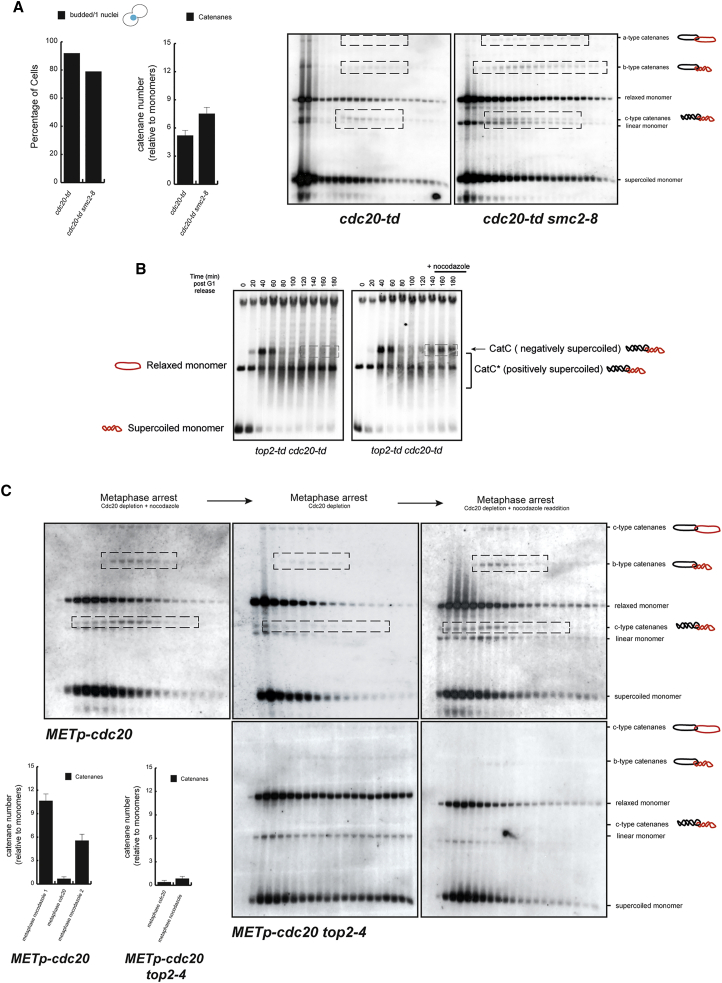
(A) cdc20-td and cdc20-td smc2-8 strains bearing the 10 kb circular minichromosome were synchronized in G1 and released into the cell cycle under conditions of Cdc20 depletion at 37°C. Samples were taken at the metaphase block. Cell extracts were separated by differential sedimentation, heat denatured in 1% SDS, and analyzed by Southern blotting to reveal plasmid species. Plasmid species included monomeric forms (OCm, Lm, and CCCm) and dimeric forms, including different SCI species (Cata-, Catb-, and Catc-type catenanes). Running position for SCIs in the blots is indicated (dashed boxes). The mitotic arrest was stable during the experiment. SCIs were increased in the cdc20-td smc2-8 strain. SCIs (including a-, b-, and c-type catenanes) were quantified using ImageJ software (bottom right-hand graph; showing the mean ± SD from three independent experiments).
(B) A cdc20-td top2-td strain bearing the centromeric plasmid pRS316 was synchronized in G1, split in two, and released into the cell cycle under conditions of Cdc20 and Top2 depletion. Nocodazole was added to one of the samples after 120 min. Samples were collected every 20 min and DNA was resolved in agarose gels and probed for the circular minichromosome. Electrophoretic mobilities of monomers (OCm, Lm, and CCCm) are indicated. Dimeric forms for CatC-type catenanes (negatively supercoiled) and CatC∗-type catenanes (positively supercoiled) (Baxter et al., 2011) are also indicated. Addition of nocodazole led to the shift back from CatC∗ to CatC.
(C) A METp-CDC20 strain (Cdc20 under Methionine promoter) (top panel) bearing the 10 kb circular minichromosome was synchronized in G1 and released into the cell cycle in the presence of nocodazole under Cdc20-depleting conditions (YPD + 5 mM methionine) at 36°C. Following metaphase arrest, cells were transferred to new media lacking nocodazole but maintaining the Cdc20 depletion (hence allowing formation of mitotic spindles) for 120 min before readdition of nocodazole. Samples were taken for analysis at the first nocodazole block (metaphase arrest − Cdc20 depletion + nocodazole), 120 min after nocodazole removal (metaphase arrest − Cdc20 depletion) and 120 min after nocodazole readdition (metaphase arrest − Cdc20 depletion + nocodazole readdition). A METp-CDC20 top2-4 (bottom panel) bearing the 10 kb circular minichromosome was synchronized in G1 and released into the cell cycle under conditions of Cdc20 depletion (YPD + 5 mM methionine) at 25°C. Nocodazole was added to the culture, and the temperature was shifted to 36°C to inactivate Top2. Samples were taken at Cdc20 block and 120 min after nocodazole addition. Cell extracts were separated by differential sedimentation, heat denatured in 1% SDS, and analyzed by Southern blotting to reveal plasmid species. Plasmid species included monomeric (OCm, Lm, and CCCm) and dimeric forms, including different SCI species (Catc-type catenanes and Catb-type catenanes). Running position for SCIs in the blots is indicated (dashed boxes). The mitotic arrest was stable during the experiments. SCIs (including c- and b-type catenanes) were quantified using ImageJ software (bottom left-hand graph; showing the mean ± SD from three independent experiments). SCIs were reduced when nocodazole was removed, and reappeared upon its readdition in the presence of Top2, but not in its absence.