Figure 1.
Mutations Identified in GLDN in AMC-Affected Families
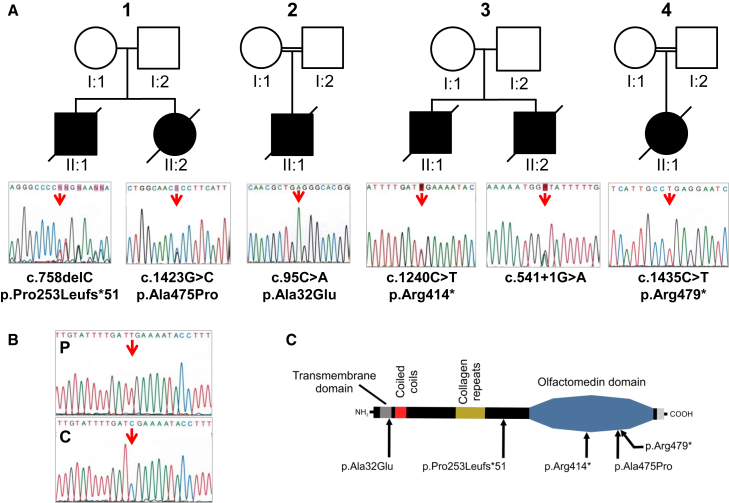
(A) Pedigrees for families 1, 2, 3, and 4 are shown. Arrows indicate mutant nucleotide positions. The affected individuals carry either compound heterozygous (families 1 and 3) or homozygous (families 2 and 4) mutations. The nucleotide and amino acid changes are indicated with respect to the GenBank: NM_181789.2 and Genbank: NP_861454.2 reference sequences, respectively. Open symbols, unaffected; filled symbols, affected.
(B) Sequencing of RT-PCR product from the RNA of the family 3 affected individual, II:1, (P) and a control individual (C) is shown. RT-PCR analysis was performed from RNA after reverse transcription with random hexamers. Long-range PCR amplification was carried out with Advantage GC Genomic LA polymerase (Clontech), then Sanger sequencing was performed from the RT-PCR products with primers GLDN-Ex1Fq and GLDN-Ex10Rq (Table S1). Sequence analysis revealed the nonsense mutation, only indicating that the allelic splice donor mutation leads to either a retention of large intronic sequence or instability of the corresponding transcript.
(C) Location of mutations relative to the predicted protein domains of gliomedin is shown.