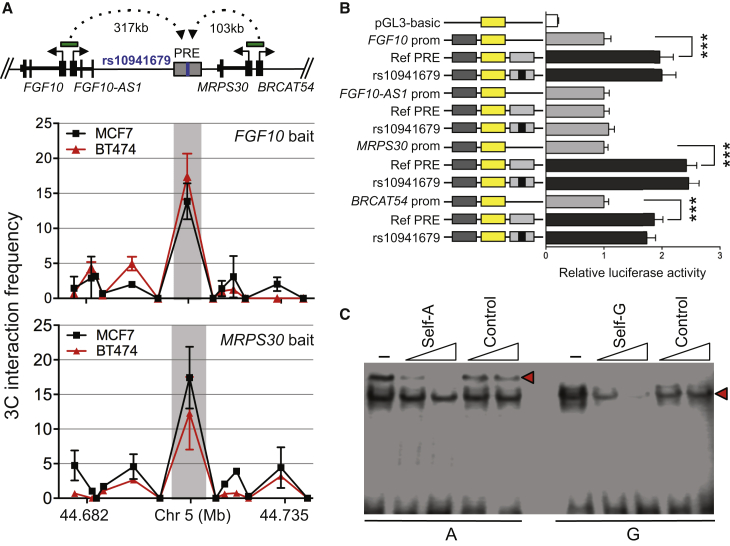
Figure 3.
Distal Regulation of FGF10 and MRPS30 at the 5p12 Risk Region
(A) 3C interaction profiles between the FGF10/FGF10AS-1 or MRPS30/BRCAT54 bidirectional promoters and the putative regulatory element (PRE; gray bar) containing SNP rs10941679. Anchor points are set at the promoters. Graphs represent one of three independent experiments (see Figure S5B). Error bars denote SD.
(B) Luciferase reporter assays after transient transfection of ER+ BT474 breast cancer cell lines. The PRE containing the major SNP allele was cloned downstream of target gene promoter-driven luciferase constructs (Ref PRE). The risk g-allele was engineered into the constructs and designated by the rs ID. Primers are listed in Table S7. Error bars denote 95% confidence intervals from three independent experiments. p values were determined by 2-way ANOVA followed by Dunnett’s multiple comparisons test (∗∗∗p < 0.001).
(C) EMSA for oligonucleotides containing SNP rs1094617 with the A = common allele and G = minor allele as indicated below the panel, assayed using BT474 nuclear extracts. Primers are listed in Table S8. Labels above each lane indicate inclusion of competitor oligonucleotides at 30- and 100-fold molar excess, respectively: (-) no competitor and control denotes a non-specific competitor. A red arrowhead shows a band of different mobility detected between the common and minor alleles.