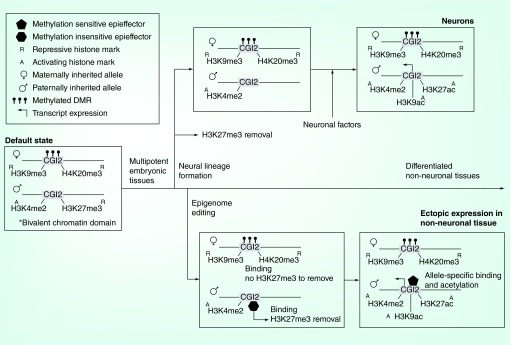
Figure 2. . Model of ectopic resolution of the Grb10 imprinting architecture by epigenome engineering.
The default state of the imprinted Grb10 CpG Island 2 locus is characterized by repressive DNA methylation and histone marks on the maternally inherited allele and a bivalent chromatin domain on the paternally inherited allele. The bivalent chromatin domain is resolved when the repressive H3K27me3 mark is lost during normal neural differentiation. Further on in differentiation, and likely as a result of neuron-specific factors, the site acquires increased H3K9 and H3K27 acetylation – both activating marks. Epigenome editing could be employed to attempt ectopic resolution of the bivalent chromatin domain and to mimic the construction of the activating architecture. In this model, a methylation insensitive EpiEffector with H3K27me3 demethylation activity targeted to the site could resolve the bivalent domain, without effect on the maternal allele. This is followed by treatment with methylation-sensitive EpiEffectors possessing H3K9 and H3K27 acetylation activity. These bind in an allele-specific manner to the unmethylated paternal allele to reconstruct a neuron-specific histone profile in non-neuronal tissues.
DMR: Differentially methylated region.