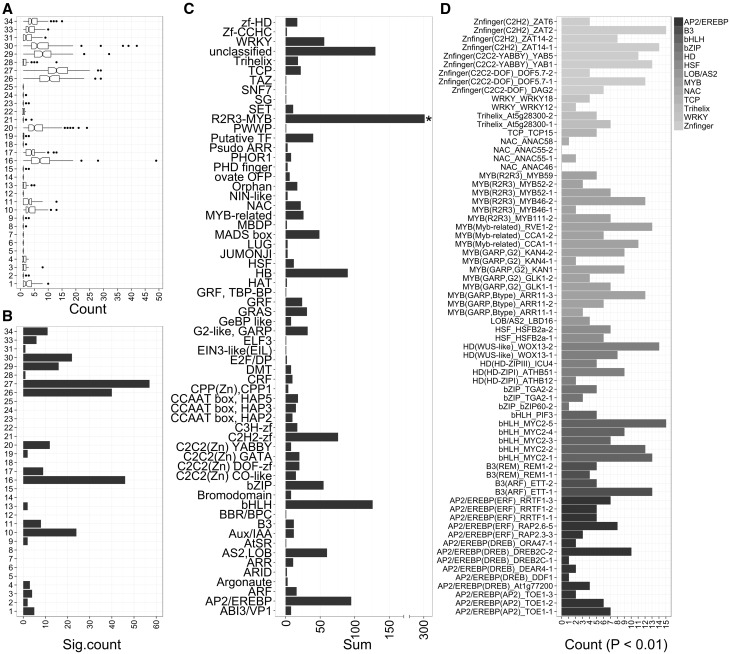
Figure 3.
Distribution of MapMan BIN terms and enriched CRE within the MYB GCN. (A) Boxplot represents the number of co-expressed genes associated with a BIN term. (B) Bar graph depicting the number of MYB GCN which are enriched (Adj. P value < 0.05) with at least one BIN term. Only high-level MapMan BIN terms were considered to provide an overview. BIN1, PS; BIN2, major CHO metabolism, BIN3, minor CHO metabolism; BIN4, glycolysis; BIN5, fermentation; BIN6, gluconeogenesis/glyoxylate cycle; BIN7, OPP; BIN8, TCA/org transformation; BIN 9, mitochondrial electron transport/ATP synthesis; BIN10, cell wall; BIN11, lipid metabolism; BIN12, N-metabolism; BIN13, amino acid metabolism; BIN13, amino acid metabolism; BIN14, S-assimilation; BIN15, metal handling; BIN16, secondary metabolism; BIN17, hormone metabolism; BIN18, co-factor and vitamin metabolism; BIN19, tetrapyrrole synthesis; BIN20, stress; BIN21, redox; BIN22, polyamine metabolism; BIN23, nucleotide metabolism; BIN24, biodegradation of xenobiotics; BIN25, C1-metabolism; BIN26, misc; BIN27, RNA; BIN28, DNA; BIN29, protein; BIN30, signalling; BIN31, cell; BIN33, development; BIN34, transport. (C) Distribution of TF family terms (BIN27.3) within the MYB GCN. Bar graph depicts the total number of genes within each TF family (sub-BIN) from the sum of MYB co-expressed genes. (D) Distribution of enriched CRE, shown as the number of MYB GCNs enriched with the respective CRE at P < 0.01. Different grey tones are depicted for each TF family to aid visualization.