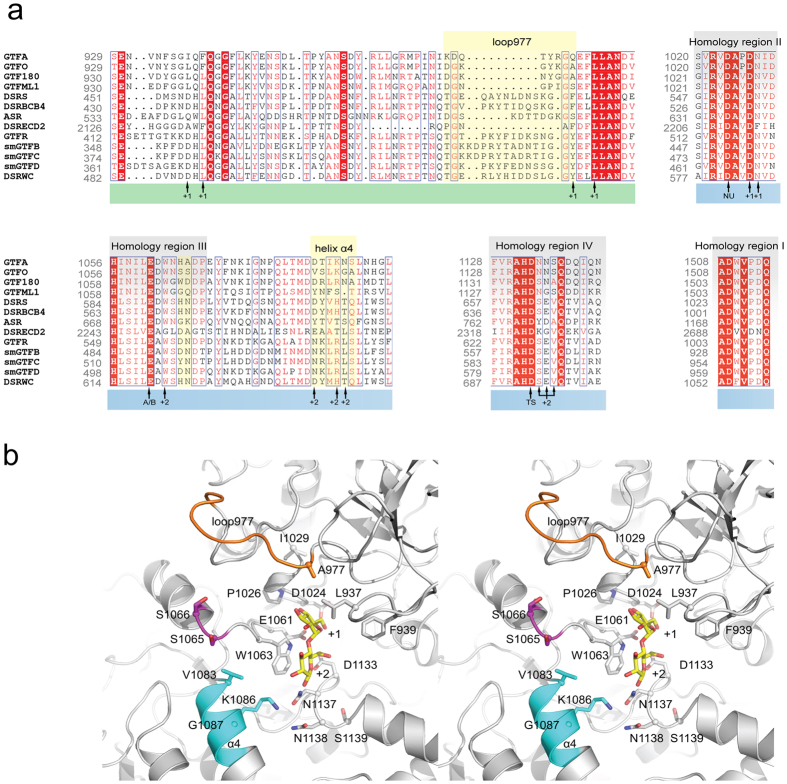
Figure 1.
(a) Partial alignment of glucansucrase amino acid sequences of family GH70. Residues from domain A are highlighted with blue at the bottom and residues from domain B are highlighted in green at the bottom. Homology regions I to IV of glucansucrases are highlighted in gray. Regions targeted for mutagenesis in this study are highlighted in yellow. Catalytic residues and residues involved in shaping acceptor binding sites +1 and +2 are indicated. (b) Stereo figure showing a model of the GTFO active site in cartoon representation, with the regions and residues targeted for mutation highlighted in color: loop977 (residues 970DGKGYKGA977, orange), S1065-S1066 (magenta) and the N-terminal part of helix α4 (residues 1083VSLKGA1088, cyan). The maltose bound in subsites +1 and +2 of L. reuteri 180 GTF180-ΔN16 is shown with yellow carbon atoms. The catalytic residues of GTFO (D1024, E1061 and D1133), and other residues surrounding the active site, are shown in stick representation.