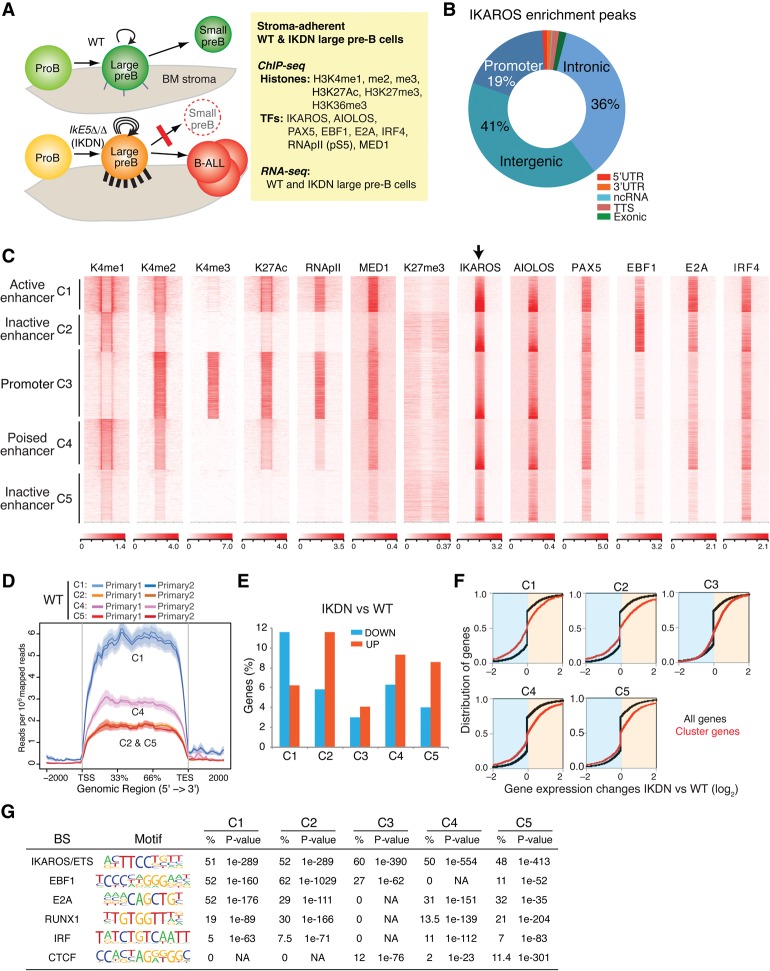
Figure 1.
Transcriptional and epigenetic landscape of wild-type adherent large pre-B cells. (A) Outline of an experimental approach to delineate the transcriptional mechanisms that guide pre-B-cell differentiation through a highly proliferative stromal-adherent phase. The effects of homozygous deletion of Ikzf1 exon 5 (IkE5Δ/Δ), encoding the DNA-binding domain and generating an IKDN isoform, on cell adhesion (cell projections on BM stroma), self-renewal (circular arrows), differentiation block (red line), and leukemogenesis (B-ALL) are depicted. (B) Relative distribution of IKAROS enrichment peaks at the indicated genomic locations in wild-type stromal-adherent large pre-B cells. (C) Heat map generated by K-means clustering of reads from ChIPs of histone modifications and transcription factors centered on IKAROS enrichment peaks (±15 kb). Read distribution within IKAROS enrichment peaks and size were normalized to the expected fragment size using the default spline fit algorithm of NGS.plot. Five clusters were generated, four of which were designated by histone modification enrichment as enhancers (C1, C2, C4, and C5) and one of which was designated as a promoter (C3). (D) Average gene expression for IKAROS gene targets in enhancer clusters C1, C2, C4, and C5, shown as exonic read distribution over the gene body (read count per million mapped reads). RNA-seq studies were performed with two independently isolated wild-type large pre-B-cell samples. (E) Percentage distribution of 1597 up-regulated and 610 down-regulated genes in IKDN pre-B cells in clusters (as in C) defined by IKAROS binding, histone modifications, and transcription factor distributions. (F) Cumulative distribution function (CDF) plots depicting differential gene expression between IKDN and wild-type pre-B cells are shown for each gene cluster (red) defined in C relative to differential gene expression of all genes (black). P-values for differences in gene expression for each cluster relative to all genes were <0.0001, calculated using a two-sided Kolmogorov Smirnov test. (G) De novo transcription factor-binding motif analysis (HOMER) of IKAROS enrichment peaks in wild-type adherent large pre-B cells is shown for each cluster defined in C. The most significantly discovered motifs, frequency (percentage), and P-values are shown.