Figure 3.
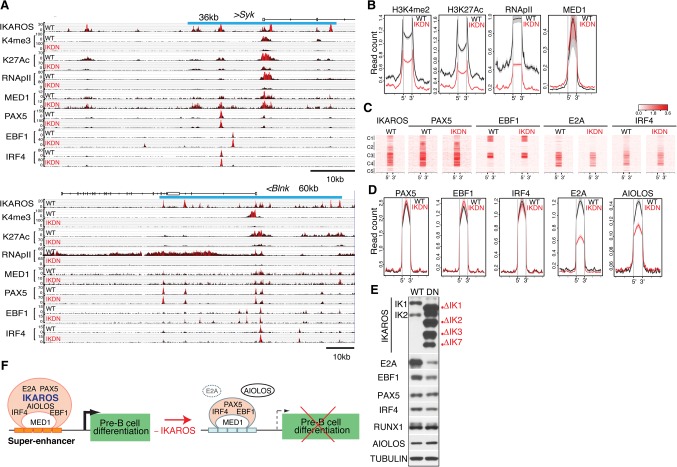
The permissive chromatin state of pre-B-cell superenhancers is dependent on IKAROS. (A) Genome browser tracks are shown for ChIP-seq of IKAROS, H3K4me3, H3K27ac, RNApIIS5, MED1, PAX5, EBF1, and IRF4 at the Sykb (top panel) and Blnk (bottom panel) genes in wild-type and IKDN pre-B cells. The associated IKAROS superenhancers are demarcated by blue lines (Sykb, 36 kb; Blnk, 60 kb). Black histograms depict sequencing read distribution, with red indicating a higher read depth. (B) Comparative analysis of read densities for permissive histone modifications (H3K27me2, H3K27ac, RNApII, and MED1) at transcription factor-binding sites that constitute superenhancers affiliated with down-regulated genes is shown in wild-type (black) and IKDN (red) pre-B cells. (C,D) Comparative analysis of B-cell transcription factor enrichment at superenhancer-binding sites as described in B. (C) Heat maps were generated by K-means clustering of reads from ChIPs for transcription factors centered at the constituent binding sites of superenhancers associated with 127 down-regulated genes described in Figure 2B. (D) Read densities of transcription factors at superenhancer-binding sites as in B and C (±15 kb). (E) Immunoblot analysis of B-cell transcription factors in wild-type and IKDN stromal-adherent large pre-B cells. The two major IKAROS isoforms (IK1 and IK2) expressed in wild-type pre-B cells are identified at the left. Mutant isoforms are identified at the right. (F) A model of regulation of pre-B-cell differentiation genes by IKAROS superenhancers. Transcriptionally permissive IKAROS-based superenhancers (orange bars) are associated with highly expressed genes promoting pre-B-cell differentiation (black arrow). Loss of IKAROS causes restriction in the chromatin configuration at superenhancers (light-blue bars), transcription is attenuated (broken arrow), and pre-B-cell differentiation is blocked. However, with the exception of AIOLOS and E2A, binding of PAX5, EBF1, IRF4, and MED1 at these regulatory sites was not affected.