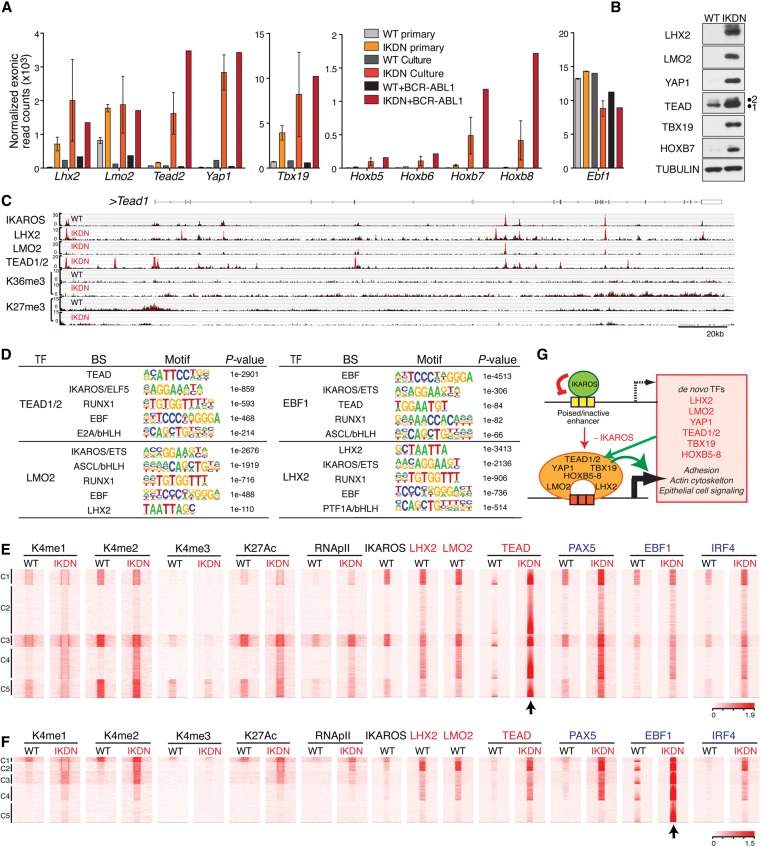
Figure 4.
Direct repression of an extralineage transcription factor network by IKAROS. (A) Induction of mRNA expression for Lhx2, Lmo2, Tead2, Yap1, Tbx19, and Hoxb5–8 in IKDN preleukemic and leukemic (+BCR-ABL1) pre-B cells. Normalized exonic raw reads are shown for the average of two wild-type and two IKDN ex vivo isolated large pre-B-cell populations (primary), one wild-type and two IKDN cultured stromal-adherent large pre-B-cell populations (Culture), one wild-type + BCR–ABL1 and one IKDN + BCR–ABL1 cultured large pre-B leukemia cell population. Ebf1 expression is shown as a control. (B) Immunoblot analysis of protein expression for the transcription factors from A. (C) Genome browser tracks are shown at the Tead1 locus for ChIP-seq of IKAROS, LHX2, LMO2, TEAD 1/2, H3K36me3 (K36me3), and H3K27me3 (K27me3) in wild-type and IKDN pre-B cells. (D) De novo motif discovery (HOMER) for peaks of TEAD (1/2), EBF1, LHX2, and LMO2 specifically enriched in IKDN stromal-adherent large pre-B cells. The most significant discovered motifs and log10 P-values are shown. (E,F) Heat maps were generated by K-means clustering of reads from ChIPs for histone modifications (H3K4me1, H3K4me2, H3K4me3, and H3K27ac) and transcription factors (RNApII, IKAROS, LHX2, LMO2, TEAD, PAX5, EBF1, and IRF4) centered (arrow) at de novo TEAD (E) or EBF1 (F) peaks (C1–C5). Extralineage and B-cell transcription factors are marked in red and blue, respectively. (G) Model of IKAROS repression at inactive/poised enhancers associated with master transcription regulators of stem and epithelial cell origin. Induction of these transcription factors triggered by loss of IKAROS initiates a feed-forward cross-regulatory loop that augments their own expression and that of other genes involved in epithelial cell functions.