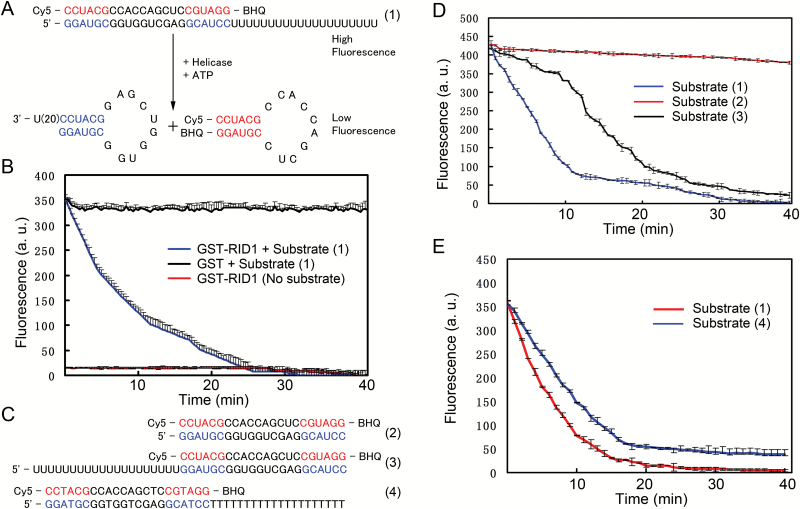
Fig. 2.
Helicase activity of RID1 unwinding dsRNA/DNA according to the molecular beacon helicase assay (MBHA). (A) The fluorescent strand in the 3′ overhanging RNA substrate (1) containing a 20-bp hanging tail was modified with Cy5 at the 3′ end and BHQ at the 5′ end. The fluorescence of Cy5 was quenched after the substrate RNA double strands were unwound and the fluorescent strand formed a stem-loop structure, which placed Cy5 near BHQ. (B) The RID1-GST fusion protein, but not GST alone, decreased the intensity of fluorescence in the MBHA system in which substrate (1) was used. A reaction system lacking RNA substrate was used as the control. a.u., arbitrary units.(C) The fluorescent strands of the blunt-ended RNA substrate (2), 5′ overhanging RNA substrate (3), and 3′ overhanging DNA substrate (4) were modified with Cy5 at the 3′ end and BHQ at the 5′ end. (D) RID1 could unwind RNA double strands from the 3′ to 5′ and 5′ to 3′ directions. The components of the MBHA system are described in the Methods section. Error bars represent the standard deviation of three independent experiments. a.u., arbitrary units. (E) RID1 could unwind RNA and DNA double strands. The components of the MBHA system are described in the Methods section. Error bars represent the standard deviation of three independent experiments. a.u., arbitrary units.