Figure 1.
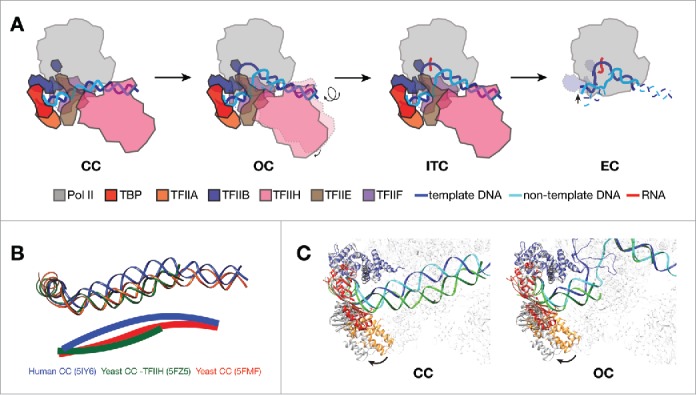
Structural features of Pol II promoter opening. (A) Schematic showing transitions from CC to EC in mammalian transcription system. PIC models are depicted as viewing down from Pol II stalk toward the active site. In CC, promoter DNA is positioned over the cleft of Pol II. The transition into OC involves the rotation and insertion of 12 bp of downstream DNA into the active center cleft by TFIIH. After loading of the template strand into the active center, Pol II PIC starts RNA synthesis and enters the ITC state. When RNA reaches a certain length, displacing the TFIIB (dotted outline in EC) zinc ribbon domain, Pol II clears promoter and turns into a processive elongation mode. (B) DNA paths in CC from three eukaryotic models,11-13 viewing from the same angle as in A. Pol II core subunits were aligned in the three CC structures for the purpose of comparison. DNA paths are also depicted as solid lines through the center of the duplex in the lower panel. Human promoter DNA is bent toward the active site, while yeast structures show clearly different bending properties of promoter DNA. (C) Comparing the positions of TFIIB-TBP-TFIIA between human (colored; PDB ID 5IY611 for CC and 5IY711 for OC) and yeast (gray; 5FZ512 for CC and 5FYW12 for OC) structures by aligning the Pol II core subunits. The TFIIB-TBP-TFIIA lobe rotates away from the rest of PIC in the yeast models. The color scheme for human structures are same as in A, except that the DNA molecule in yeast structures are shown in green (dark green for template strand, and light green for non-template strand).
