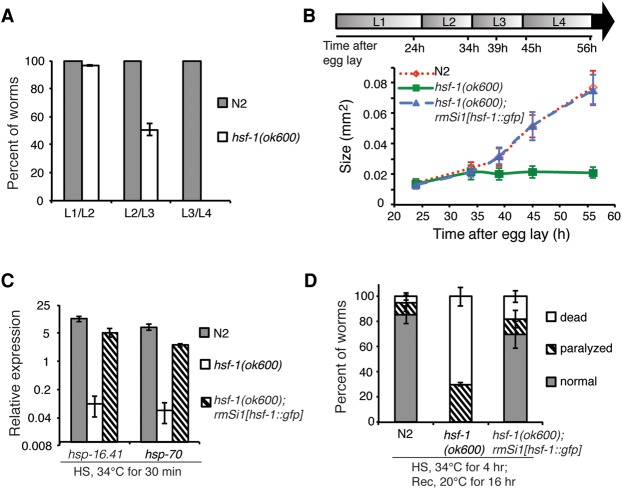
Figure 1.
HSF-1 is essential for C. elegans larval development. (A) Histograms showing the percent of wild-type (N2) and hsf-1(ok600) animals that passed through each larval molt at 20°C. Error bars represent the SEM of four biological replicates. n = 30 N2; n = 55 hsf-1(ok600). (B) The size of wild-type (N2), hsf-1(ok600), and hsf-1(ok600); rmSi1[hsf-1::gfp] larvae. The rmSi1 transgene carries a single copy of hsf-1p::hsf-1(minigene)::gfp::hsf-1 3′ UTR. At least 30 animals of each genotype were measured. Error bars represent SD. The size of hsf-1(ok600) animals is unchanged from 34 h after egg lay (L2 stage; P = 0.68, one-way ANOVA analysis) and significantly differs from N2 and hsf-1(ok600); rmSi1[hsf-1::gfp] measured at 39 h after egg lay (L3 stage) and thereafter. t-test, P = 0.001. The timeline represents larval stages (L1–L4) at given time points. (C) The expression of hsp-16.41 and hsp-70(C12C8.1) in wild-type (N2), hsf-1(ok600), and hsf-1(ok600); rmSi1[hsf-1::gfp] animals exposed to heat shock (HS; 30 min at 34°C) at L2 stage. Error bars represent the SEM of biological triplicates. (D) Thermorecovery of wild-type (N2), hsf-1(ok600), and hsf-1(ok600); rmSi1[hsf-1::gfp] animals at L2 (30 h after egg lay) exposed to an extended heat shock (HS) for 4 h at 34°C followed by recovery (Rec) for 16 h at 20°C. Error bars represent the SEM of biological triplicates. n = 137 N2; n = 130 hsf-1(ok600); n = 136 hsf-1(ok600); rmSi1[hsf-1::gfp].